A feedback loop comprising PRMT7 and miR-24-2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness
- PMID: 27625395
- PMCID: PMC5159542
- DOI: 10.1093/nar/gkw788
A feedback loop comprising PRMT7 and miR-24-2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness
Abstract
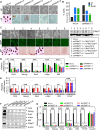
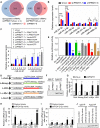
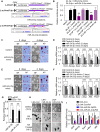
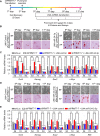
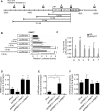
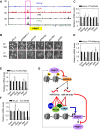
Self-renewal and pluripotency are two fundamental characteristics of embryonic stem cells (ESCs) and are controlled by diverse regulatory factors, including pluripotent factors, epigenetic regulators and microRNAs (miRNAs). Although histone methyltransferases are key epigenetic regulators, whether and how a histone methyltransferase forms a network with miRNAs and the core pluripotent factor system to regulate ESC stemness is little known. Here, we show that the protein arginine methyltransferase 7 (PRMT7) is a pluripotent factor essential for the stemness of mouse ESCs. PRMT7 repressed the miR-24-2 gene encoding miR-24-3p and miR-24-2-5p by upregulating the levels of symmetrically dimethylated H4R3. Notably, miR-24-3p targeted the 3' untranslated regions (UTRs) of the major pluripotent factors Oct4, Nanog, Klf4 and c-Myc, whereas miR-24-2-5p silenced Klf4 and c-Myc expression. miR-24-3p and miR-24-2-5p also targeted the 3'UTR of their repressor gene Prmt7 miR-24-3p and miR-24-2-5p induced mouse ESC differentiation, and their anti-sense inhibitors substantially reversed spontaneous differentiation of PRMT7-depleted mouse ESCs. Oct4, Nanog, Klf4 and c-Myc positively regulated Prmt7 expression. These findings define miR-24-3p and miR-24-2-5p as new anti-pluripotent miRNAs and also reveal a novel epigenetic stemness-regulatory mechanism in which a double-negative feedback loop consisting of PRMT7 and miR-24-3p/miR24-2-5p interplays with Oct4, Nanog, Klf4 and c-Myc to control ESC stemness.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Protein arginine methyltransferase 7-mediated microRNA-221 repression maintains Oct4, Nanog, and Sox2 levels in mouse embryonic stem cells.J Biol Chem. 2018 Mar 16;293(11):3925-3936. doi: 10.1074/jbc.RA117.000425. Epub 2018 Jan 29. J Biol Chem. 2018. PMID: 29378844 Free PMC article.
-
CHIR99021 enhances Klf4 Expression through β-Catenin Signaling and miR-7a Regulation in J1 Mouse Embryonic Stem Cells.PLoS One. 2016 Mar 3;11(3):e0150936. doi: 10.1371/journal.pone.0150936. eCollection 2016. PLoS One. 2016. PMID: 26938105 Free PMC article.
-
H3K4 Methyltransferase Set1a Is A Key Oct4 Coactivator Essential for Generation of Oct4 Positive Inner Cell Mass.Stem Cells. 2016 Mar;34(3):565-80. doi: 10.1002/stem.2250. Epub 2016 Jan 19. Stem Cells. 2016. PMID: 26785054
-
Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review.
-
Subcellular localisation of the stem cell markers OCT4, SOX2, NANOG, KLF4 and c-MYC in cancer: a review.J Clin Pathol. 2018 Jan;71(1):88-91. doi: 10.1136/jclinpath-2017-204815. Epub 2017 Nov 27. J Clin Pathol. 2018. PMID: 29180509 Review.
Cited by
-
Recent Advances in Understanding Cholangiocarcinoma.F1000Res. 2017 Oct 9;6:1818. doi: 10.12688/f1000research.12118.1. eCollection 2017. F1000Res. 2017. PMID: 29067165 Free PMC article. Review.
-
Asymmetrical methyltransferase PRMT3 regulates human mesenchymal stem cell osteogenesis via miR-3648.Cell Death Dis. 2019 Aug 5;10(8):581. doi: 10.1038/s41419-019-1815-7. Cell Death Dis. 2019. PMID: 31378783 Free PMC article.
-
Proteomic analysis of the IPF mesenchymal progenitor cell nuclear proteome identifies abnormalities in key nodal proteins that underlie their fibrogenic phenotype.Proteomics. 2022 Jul;22(13-14):e2200018. doi: 10.1002/pmic.202200018. Epub 2022 Jun 14. Proteomics. 2022. PMID: 35633524 Free PMC article.
-
Inborn-like errors of metabolism are determinants of breast cancer risk, clinical response and survival: a study of human biochemical individuality.Oncotarget. 2018 Aug 3;9(60):31664-31681. doi: 10.18632/oncotarget.25839. eCollection 2018 Aug 3. Oncotarget. 2018. PMID: 30167086 Free PMC article.
-
microRNA-Mediated Encoding and Decoding of Time-Dependent Signals in Tumorigenesis.Biomolecules. 2022 Jan 26;12(2):213. doi: 10.3390/biom12020213. Biomolecules. 2022. PMID: 35204714 Free PMC article. Review.
References
-
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. - PubMed
-
- Spivakov M., Fisher A.G. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007;8:263–271. - PubMed
-
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

