ChIA-PET2: a versatile and flexible pipeline for ChIA-PET data analysis
- PMID: 27625391
- PMCID: PMC5224499
- DOI: 10.1093/nar/gkw809
ChIA-PET2: a versatile and flexible pipeline for ChIA-PET data analysis
Abstract
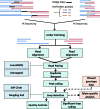
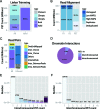
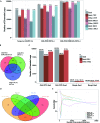
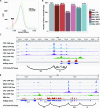
ChIA-PET2 is a versatile and flexible pipeline for analyzing different types of ChIA-PET data from raw sequencing reads to chromatin loops. ChIA-PET2 integrates all steps required for ChIA-PET data analysis, including linker trimming, read alignment, duplicate removal, peak calling and chromatin loop calling. It supports different kinds of ChIA-PET data generated from different ChIA-PET protocols and also provides quality controls for different steps of ChIA-PET analysis. In addition, ChIA-PET2 can use phased genotype data to call allele-specific chromatin interactions. We applied ChIA-PET2 to different ChIA-PET datasets, demonstrating its significantly improved performance as well as its ability to easily process ChIA-PET raw data. ChIA-PET2 is available at https://github.com/GuipengLi/ChIA-PET2.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Mango: a bias-correcting ChIA-PET analysis pipeline.Bioinformatics. 2015 Oct 1;31(19):3092-8. doi: 10.1093/bioinformatics/btv336. Epub 2015 Jun 1. Bioinformatics. 2015. PMID: 26034063 Free PMC article.
-
ChIAPoP: a new tool for ChIA-PET data analysis.Nucleic Acids Res. 2019 Apr 23;47(7):e37. doi: 10.1093/nar/gkz062. Nucleic Acids Res. 2019. PMID: 30753588 Free PMC article.
-
Chromatin Interaction Analysis with Updated ChIA-PET Tool (V3).Genes (Basel). 2019 Jul 22;10(7):554. doi: 10.3390/genes10070554. Genes (Basel). 2019. PMID: 31336684 Free PMC article.
-
Methods for comparative ChIA-PET and Hi-C data analysis.Methods. 2020 Jan 1;170:69-74. doi: 10.1016/j.ymeth.2019.09.019. Epub 2019 Oct 16. Methods. 2020. PMID: 31629084 Review.
-
Progresses in the plant 3D chromatin architecture.Yi Chuan. 2020 Jan 20;42(1):73-86. doi: 10.16288/j.yczz.19-326. Yi Chuan. 2020. PMID: 31956098 Review.
Cited by
-
3D genome architecture coordinates trans and cis regulation of differentially expressed ear and tassel genes in maize.Genome Biol. 2020 Jun 16;21(1):143. doi: 10.1186/s13059-020-02063-7. Genome Biol. 2020. PMID: 32546248 Free PMC article.
-
3DeFDR: statistical methods for identifying cell type-specific looping interactions in 5C and Hi-C data.Genome Biol. 2020 Aug 28;21(1):219. doi: 10.1186/s13059-020-02061-9. Genome Biol. 2020. PMID: 32859248 Free PMC article.
-
YY1 safeguard multidimensional epigenetic landscape associated with extended pluripotency.Nucleic Acids Res. 2022 Nov 28;50(21):12019-12038. doi: 10.1093/nar/gkac230. Nucleic Acids Res. 2022. PMID: 35425987 Free PMC article.
-
CTCF organizes inter-A compartment interactions through RYBP-dependent phase separation.Cell Res. 2022 Aug;32(8):744-760. doi: 10.1038/s41422-022-00676-0. Epub 2022 Jun 29. Cell Res. 2022. PMID: 35768498 Free PMC article.
-
An alternative CTCF isoform antagonizes canonical CTCF occupancy and changes chromatin architecture to promote apoptosis.Nat Commun. 2019 Apr 4;10(1):1535. doi: 10.1038/s41467-019-08949-w. Nat Commun. 2019. PMID: 30948729 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

