Effects of Folic Acid on Secretases Involved in Aβ Deposition in APP/PS1 Mice
- PMID: 27618097
- PMCID: PMC5037541
- DOI: 10.3390/nu8090556
Effects of Folic Acid on Secretases Involved in Aβ Deposition in APP/PS1 Mice
Abstract
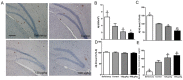
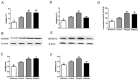
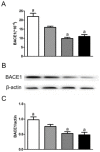
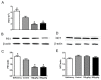
Alzheimer's disease (AD) is the most common type of dementia. Amyloid-β protein (Aβ) is identified as the core protein of neuritic plaques. Aβ is generated by the sequential cleavage of the amyloid precursor protein (APP) via the APP cleaving enzyme (α-secretase, or β-secretase) and γ-secretase. Previous studies indicated that folate deficiency elevated Aβ deposition in APP/PS1 mice, and this rise was prevented by folic acid. In the present study, we aimed to investigate whether folic acid could influence the generation of Aβ by regulating α-, β-, and γ-secretase. Herein, we demonstrated that folic acid reduced the deposition of Aβ42 in APP/PS1 mice brain by decreasing the mRNA and protein expressions of β-secretase [beta-site APP-cleaving enzyme 1 (BACE1)] and γ-secretase complex catalytic component-presenilin 1 (PS1)-in APP/PS1 mice brain. Meanwhile, folic acid increased the levels of ADAM9 and ADAM10, which are important α-secretases in ADAM (a disintegrin and metalloprotease) family. However, folic acid has no impact on the protein expression of nicastrin (Nct), another component of γ-secretase complex. Moreover, folic acid regulated the expression of miR-126-3p and miR-339-5p, which target ADAM9 and BACE1, respectively. Taken together, the effect of folic acid on Aβ deposition may relate to making APP metabolism through non-amyloidogenic pathway by decreasing β-secretase and increasing α-secretase. MicroRNA (miRNA) may involve in the regulation mechanism of folic acid on secretase expression.
Keywords: Alzheimer’s disease; Aβ generation; folic acid; microRNAs; secretase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Osmundacetone ameliorates Alzheimer's-like pathologies by inhibiting β-amyloid fibrillation, oxidative damage and neuroinflammation in APP/PS1 transgenic mice.Phytomedicine. 2024 Dec;135:156091. doi: 10.1016/j.phymed.2024.156091. Epub 2024 Sep 21. Phytomedicine. 2024. PMID: 39332101
-
1950 MHz Electromagnetic Fields Ameliorate Aβ Pathology in Alzheimer's Disease Mice.Curr Alzheimer Res. 2015;12(5):481-92. doi: 10.2174/156720501205150526114448. Curr Alzheimer Res. 2015. PMID: 26017559 Free PMC article.
-
Farnesyltransferase inhibitor LNK-754 attenuates axonal dystrophy and reduces amyloid pathology in mice.Mol Neurodegener. 2022 Aug 20;17(1):54. doi: 10.1186/s13024-022-00561-9. Mol Neurodegener. 2022. PMID: 35987691 Free PMC article.
-
Cathepsin B Deficiency Improves Memory Deficits and Reduces Amyloid-β in hAβPP Mouse Models Representing the Major Sporadic Alzheimer's Disease Condition.J Alzheimers Dis. 2023;93(1):33-46. doi: 10.3233/JAD-221005. J Alzheimers Dis. 2023. PMID: 36970896 Free PMC article. Review.
-
APP Function and Lipids: A Bidirectional Link.Front Mol Neurosci. 2017 Mar 10;10:63. doi: 10.3389/fnmol.2017.00063. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28344547 Free PMC article. Review.
Cited by
-
The Use of Bioactive Compounds in Hyperglycemia- and Amyloid Fibrils-Induced Toxicity in Type 2 Diabetes and Alzheimer's Disease.Pharmaceutics. 2022 Jan 20;14(2):235. doi: 10.3390/pharmaceutics14020235. Pharmaceutics. 2022. PMID: 35213966 Free PMC article. Review.
-
Cognitive Function After Stopping Folic Acid and DHA Intervention: An Extended Follow-Up Results from the Randomized, Double Blind, Placebo-Controlled Trial in Older Adults with Mild Cognitive Impairment.J Alzheimers Dis Rep. 2024 Sep 27;8(1):1285-1295. doi: 10.3233/ADR-240033. eCollection 2024. J Alzheimers Dis Rep. 2024. PMID: 39434820 Free PMC article.
-
Epigenetic regulation of synaptic disorder in Alzheimer's disease.Front Neurosci. 2022 Aug 3;16:888014. doi: 10.3389/fnins.2022.888014. eCollection 2022. Front Neurosci. 2022. PMID: 35992921 Free PMC article. Review.
-
LongShengZhi Capsule Attenuates Alzheimer-Like Pathology in APP/PS1 Double Transgenic Mice by Reducing Neuronal Oxidative Stress and Inflammation.Front Aging Neurosci. 2020 Nov 23;12:582455. doi: 10.3389/fnagi.2020.582455. eCollection 2020. Front Aging Neurosci. 2020. PMID: 33328962 Free PMC article.
-
One-Carbon Metabolism: Pulling the Strings behind Aging and Neurodegeneration.Cells. 2022 Jan 9;11(2):214. doi: 10.3390/cells11020214. Cells. 2022. PMID: 35053330 Free PMC article. Review.
References
-
- Abraham J.N., Kedracki D., Prado E., Gourmel C., Maroni P., Nardin C. Effect of the Interaction of the amyloid β (1-42) peptide with short single-stranded synthetic nucleotide sequences: Morphological characterization of the inhibition of fibrils formation and fibrils disassembly. Biomacromolecules. 2014;15:253–258. doi: 10.1021/bm501004q. - DOI - PubMed
-
- Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. USA. 1996;96:3922–3927. doi: 10.1073/pnas.96.7.3922. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

