Heterogeneous structures formed by conserved RNA sequences within the HIV reverse transcription initiation site
- PMID: 27613581
- PMCID: PMC5066621
- DOI: 10.1261/rna.056804.116
Heterogeneous structures formed by conserved RNA sequences within the HIV reverse transcription initiation site
Abstract
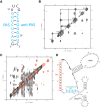
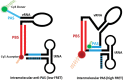
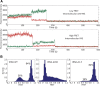
Reverse transcription is a key process in the early steps of HIV infection. This process initiates within a specific complex formed by the 5' UTR of the HIV genomic RNA (vRNA) and a host primer tRNALys3 Using nuclear magnetic resonance (NMR) spectroscopy and single-molecule fluorescence spectroscopy, we detect two distinct conformers adopted by the tRNA/vRNA initiation complex. We directly show that an interaction between the conserved 8-nucleotide viral RNA primer activation signal (PAS) and the primer tRNA occurs in one of these conformers. This intermolecular PAS interaction likely induces strain on a vRNA intramolecular helix, which must be broken for reverse transcription to initiate. We propose a mechanism by which this vRNA/tRNA conformer relieves the kinetic block formed by the vRNA intramolecular helix to initiate reverse transcription.
Keywords: HIV RNA structure; NMR spectroscopy; reverse transcription; single-molecule FRET.
© 2016 Coey et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



Similar articles
-
Secondary structure of the HIV reverse transcription initiation complex by NMR.J Mol Biol. 2011 Jul 29;410(5):863-74. doi: 10.1016/j.jmb.2011.04.024. J Mol Biol. 2011. PMID: 21763492 Free PMC article.
-
Dynamic Interplay of RNA and Protein in the Human Immunodeficiency Virus-1 Reverse Transcription Initiation Complex.J Mol Biol. 2018 Dec 7;430(24):5137-5150. doi: 10.1016/j.jmb.2018.08.029. Epub 2018 Sep 7. J Mol Biol. 2018. PMID: 30201267 Free PMC article.
-
The Interaction between tRNA(Lys) 3 and the primer activation signal deciphered by NMR spectroscopy.PLoS One. 2013 Jun 6;8(6):e64700. doi: 10.1371/journal.pone.0064700. Print 2013. PLoS One. 2013. PMID: 23762248 Free PMC article.
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
-
HIV-1 reverse transcription initiation: a potential target for novel antivirals?Virus Res. 2008 Jun;134(1-2):4-18. doi: 10.1016/j.virusres.2007.12.009. Epub 2008 Feb 6. Virus Res. 2008. PMID: 18255184 Review.
Cited by
-
The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.Nucleic Acids Res. 2021 Jun 4;49(10):5925-5942. doi: 10.1093/nar/gkab342. Nucleic Acids Res. 2021. PMID: 33978756 Free PMC article.
-
Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.J Mol Biol. 2020 Jul 24;432(16):4499-4522. doi: 10.1016/j.jmb.2020.06.003. Epub 2020 Jun 6. J Mol Biol. 2020. PMID: 32512005 Free PMC article.
-
Retroviral PBS-segment sequence and structure: Orchestrating early and late replication events.Retrovirology. 2024 Jun 17;21(1):12. doi: 10.1186/s12977-024-00646-x. Retrovirology. 2024. PMID: 38886829 Free PMC article. Review.
-
Illuminating the virus life cycle with single-molecule FRET imaging.Adv Virus Res. 2019;105:239-273. doi: 10.1016/bs.aivir.2019.07.004. Epub 2019 Aug 20. Adv Virus Res. 2019. PMID: 31522706 Free PMC article.
-
High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.Nat Commun. 2021 May 4;12(1):2500. doi: 10.1038/s41467-021-22628-9. Nat Commun. 2021. PMID: 33947853 Free PMC article.
References
-
- Baltimore D. 1970. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226: 1209–1211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
