Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice
- PMID: 27608668
- PMCID: PMC5404394
- DOI: 10.1126/science.aah3945
Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice
Abstract
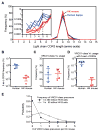
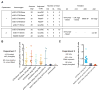
A major obstacle to a broadly neutralizing antibody (bnAb)-based HIV vaccine is the activation of appropriate B cell precursors. Germline-targeting immunogens must be capable of priming rare bnAb precursors in the physiological setting. We tested the ability of the VRC01-class bnAb germline-targeting immunogen eOD-GT8 60mer (60-subunit self-assembling nanoparticle) to activate appropriate precursors in mice transgenic for human immunoglobulin (Ig) loci. Despite an average frequency of, at most, about one VRC01-class precursor per mouse, we found that at least 29% of singly immunized mice produced a VRC01-class memory response, suggesting that priming generally succeeded when at least one precursor was present. The results demonstrate the feasibility of using germline targeting to prime specific and exceedingly rare bnAb-precursor B cells within a humanlike repertoire.
Copyright © 2016, American Association for the Advancement of Science.
Figures

Similar articles
-
HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.Science. 2016 Mar 25;351(6280):1458-63. doi: 10.1126/science.aad9195. Science. 2016. PMID: 27013733 Free PMC article.
-
B cells expressing authentic naive human VRC01-class BCRs can be recruited to germinal centers and affinity mature in multiple independent mouse models.Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):22920-22931. doi: 10.1073/pnas.2004489117. Epub 2020 Sep 1. Proc Natl Acad Sci U S A. 2020. PMID: 32873644 Free PMC article.
-
HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen.Science. 2015 Jul 10;349(6244):156-61. doi: 10.1126/science.aac5894. Epub 2015 Jun 18. Science. 2015. PMID: 26089355 Free PMC article.
-
Targeting broadly neutralizing antibody precursors: a naïve approach to vaccine design.Curr Opin HIV AIDS. 2019 Jul;14(4):294-301. doi: 10.1097/COH.0000000000000548. Curr Opin HIV AIDS. 2019. PMID: 30946041 Review.
-
Human Ig knockin mice to study the development and regulation of HIV-1 broadly neutralizing antibodies.Immunol Rev. 2017 Jan;275(1):89-107. doi: 10.1111/imr.12505. Immunol Rev. 2017. PMID: 28133799 Free PMC article. Review.
Cited by
-
Advancing mRNA technologies for therapies and vaccines: An African context.Front Immunol. 2022 Oct 24;13:1018961. doi: 10.3389/fimmu.2022.1018961. eCollection 2022. Front Immunol. 2022. PMID: 36353641 Free PMC article. Review.
-
Impact of early life exposure to ionizing radiation on influenza vaccine response in an elderly Japanese cohort.Vaccine. 2018 Oct 29;36(45):6650-6659. doi: 10.1016/j.vaccine.2018.09.054. Epub 2018 Sep 28. Vaccine. 2018. PMID: 30274868 Free PMC article.
-
Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.Nat Commun. 2022 Aug 3;13(1):4515. doi: 10.1038/s41467-022-32208-0. Nat Commun. 2022. PMID: 35922441 Free PMC article.
-
Characterisation of the immune repertoire of a humanised transgenic mouse through immunophenotyping and high-throughput sequencing.Elife. 2023 Mar 27;12:e81629. doi: 10.7554/eLife.81629. Elife. 2023. PMID: 36971345 Free PMC article.
-
Employing Broadly Neutralizing Antibodies as a Human Immunodeficiency Virus Prophylactic & Therapeutic Application.Front Immunol. 2021 Jul 20;12:697683. doi: 10.3389/fimmu.2021.697683. eCollection 2021. Front Immunol. 2021. PMID: 34354709 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

