Canonical and Variant Forms of Histone H3 Are Deposited onto the Human Cytomegalovirus Genome during Lytic and Latent Infections
- PMID: 27605676
- PMCID: PMC5105665
- DOI: 10.1128/JVI.01220-16
Canonical and Variant Forms of Histone H3 Are Deposited onto the Human Cytomegalovirus Genome during Lytic and Latent Infections
Abstract
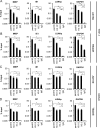
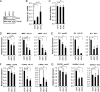
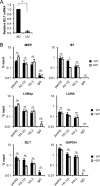
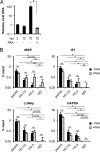
Chromatin is the nucleoprotein complex that protects and compacts eukaryotic genomes. It is responsible for a large part of the epigenetic control of transcription. The genomes of DNA viruses such as human cytomegalovirus (HCMV) are devoid of histones within virions but are chromatinized and epigenetically regulated following delivery to the host cell nucleus. How chromatin is initially assembled on viral genomes and which variant forms of the core histone proteins are deposited are incompletely understood. We monitored the deposition of both ectopically expressed and endogenous histones H3.1 and H3.2 (collectively, H3.1/2) and H3.3 during lytic and latent HCMV infections. Here, we show that they are deposited on HCMV genomes during lytic and latent infections, suggesting similar mechanisms of viral chromatin assembly during the different infection types and indicating that both canonical and variant core histones may be important modulators of infecting viral genomes. We further show that association of both H3.1/2 and H3.3 occurs independent of viral DNA synthesis or de novo viral gene expression, implicating cellular factors and/or virion components in the formation of chromatin on virion-delivered genomes during both lytic and latent infections. IMPORTANCE It is well established that infecting herpesvirus genomes are chromatinized upon entry into the host cell nucleus. Why or how this occurs is a mystery. It is important to know why they are chromatinized in order to better understand cellular pathogen recognition (DNA sensing) pathways and viral fate determinations (lytic or latent) and to anticipate how artificially modulating chromatinization may impact viral infections. It is important to know how the genomes are chromatinized in order to potentially modulate the process for therapeutic effect. Our work showing that HCMV genomes are loaded with canonical and variant H3 histones during both lytic and latent infections strengthens the hypothesis that chromatinization pathways are similar between the two infection types, implicates virion or cellular factors in this process, and exposes the possibility that histone variants, in addition to posttranslational modification, may impact viral gene expression. These revelations are important to understanding and intelligently intervening in herpesvirus infections.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
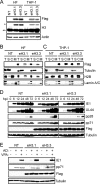
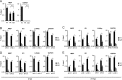
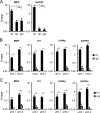
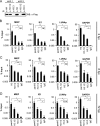
Similar articles
-
Phylogenetic analysis of the core histone doublet and DNA topo II genes of Marseilleviridae: evidence of proto-eukaryotic provenance.Epigenetics Chromatin. 2017 Nov 28;10(1):55. doi: 10.1186/s13072-017-0162-0. Epigenetics Chromatin. 2017. PMID: 29179736 Free PMC article.
-
Histone H2A variant H2A.B is enriched in transcriptionally active HSV-1 lytic chromatin.bioRxiv [Preprint]. 2023 Dec 22:2023.12.22.573075. doi: 10.1101/2023.12.22.573075. bioRxiv. 2023. Update in: J Virol. 2024 Apr 16;98(4):e0201523. doi: 10.1128/jvi.02015-23 PMID: 38187672 Free PMC article. Updated. Preprint.
-
The differential mobilization of histones H3.1 and H3.3 by herpes simplex virus 1 relates histone dynamics to the assembly of viral chromatin.PLoS Pathog. 2013;9(10):e1003695. doi: 10.1371/journal.ppat.1003695. Epub 2013 Oct 10. PLoS Pathog. 2013. PMID: 24130491 Free PMC article.
-
How to control an infectious bead string: nucleosome-based regulation and targeting of herpesvirus chromatin.Rev Med Virol. 2011 May;21(3):154-80. doi: 10.1002/rmv.690. Rev Med Virol. 2011. PMID: 21538665 Review.
-
Chromatin control of human cytomegalovirus infection.mBio. 2023 Aug 31;14(4):e0032623. doi: 10.1128/mbio.00326-23. Epub 2023 Jul 13. mBio. 2023. PMID: 37439556 Free PMC article. Review.
Cited by
-
ATRX limits the accessibility of histone H3-occupied HSV genomes during lytic infection.PLoS Pathog. 2021 Apr 28;17(4):e1009567. doi: 10.1371/journal.ppat.1009567. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33909709 Free PMC article.
-
De Novo Polycomb Recruitment: Lessons from Latent Herpesviruses.Viruses. 2021 Jul 27;13(8):1470. doi: 10.3390/v13081470. Viruses. 2021. PMID: 34452335 Free PMC article. Review.
-
Early Nuclear Events after Herpesviral Infection.J Clin Med. 2019 Sep 7;8(9):1408. doi: 10.3390/jcm8091408. J Clin Med. 2019. PMID: 31500286 Free PMC article. Review.
-
The Role of Promyelocytic Leukemia Nuclear Bodies During HPV Infection.Front Cell Infect Microbiol. 2020 Feb 19;10:35. doi: 10.3389/fcimb.2020.00035. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32154186 Free PMC article. Review.
-
Human Cytomegalovirus Genomes Survive Mitosis via the IE19 Chromatin-Tethering Domain.mBio. 2020 Sep 29;11(5):e02410-20. doi: 10.1128/mBio.02410-20. mBio. 2020. PMID: 32994332 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

