The cnidarian Hydractinia echinata employs canonical and highly adapted histones to pack its DNA
- PMID: 27602058
- PMCID: PMC5011920
- DOI: 10.1186/s13072-016-0085-1
The cnidarian Hydractinia echinata employs canonical and highly adapted histones to pack its DNA
Abstract
Background: Cnidarians are a group of early branching animals including corals, jellyfish and hydroids that are renowned for their high regenerative ability, growth plasticity and longevity. Because cnidarian genomes are conventional in terms of protein-coding genes, their remarkable features are likely a consequence of epigenetic regulation. To facilitate epigenetics research in cnidarians, we analysed the histone complement of the cnidarian model organism Hydractinia echinata using phylogenomics, proteomics, transcriptomics and mRNA in situ hybridisations.
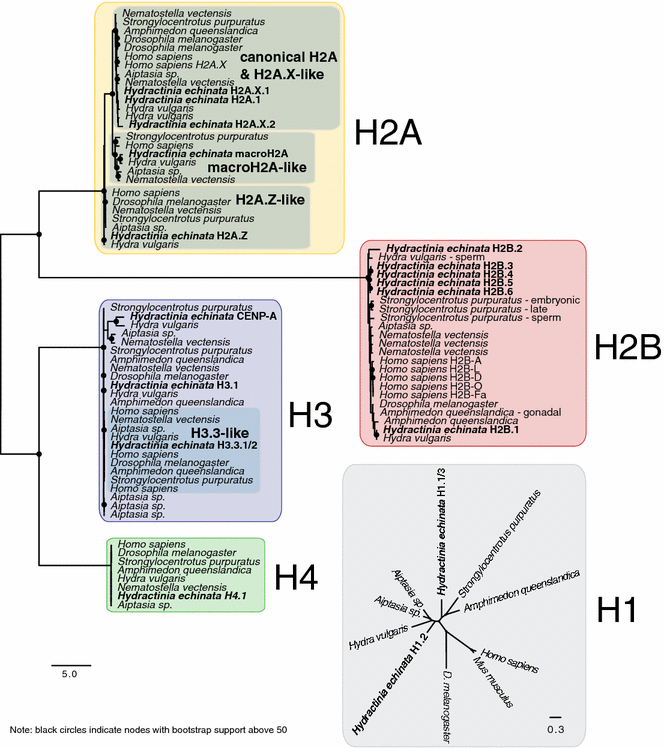
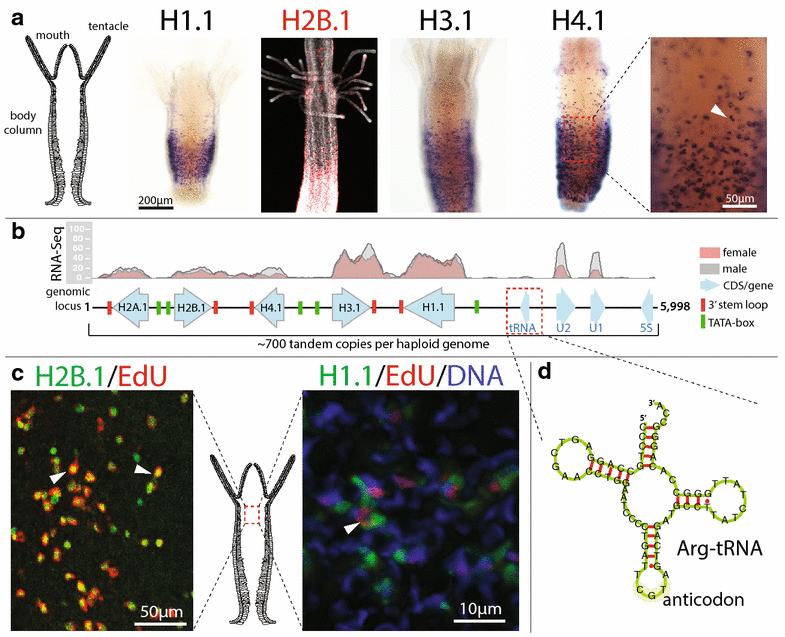
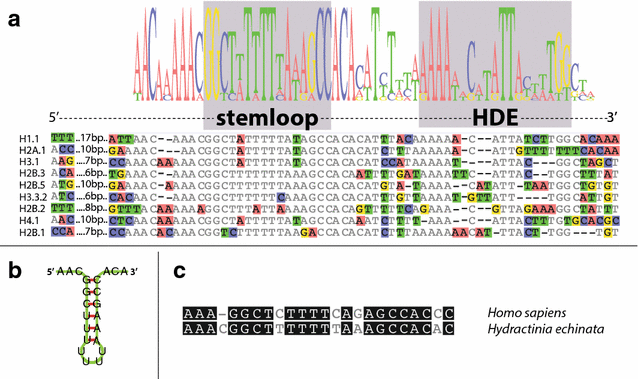
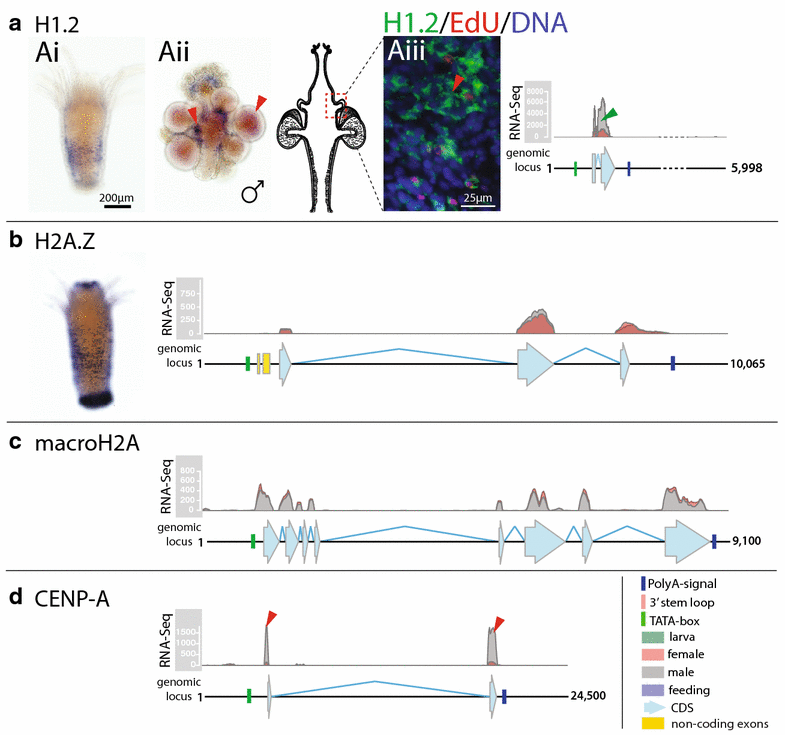
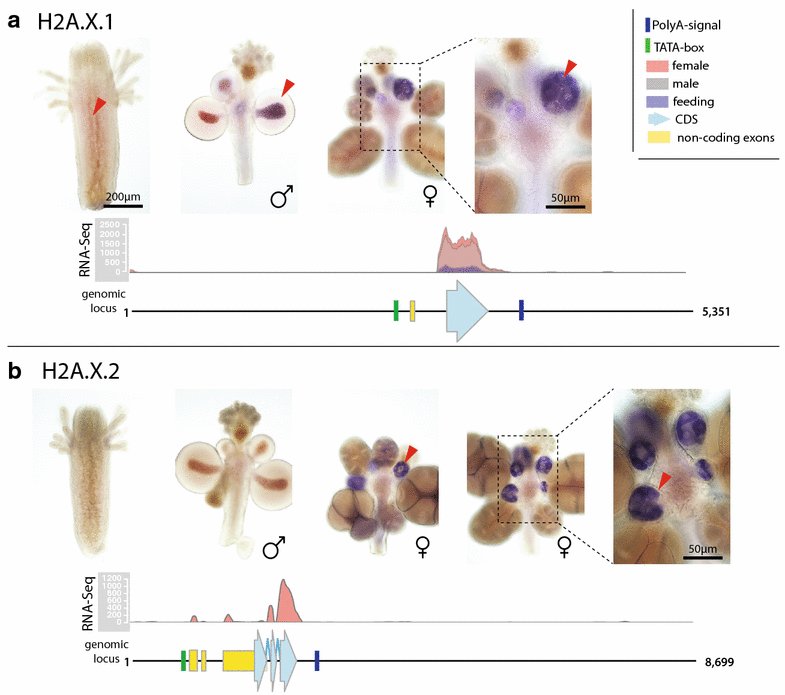
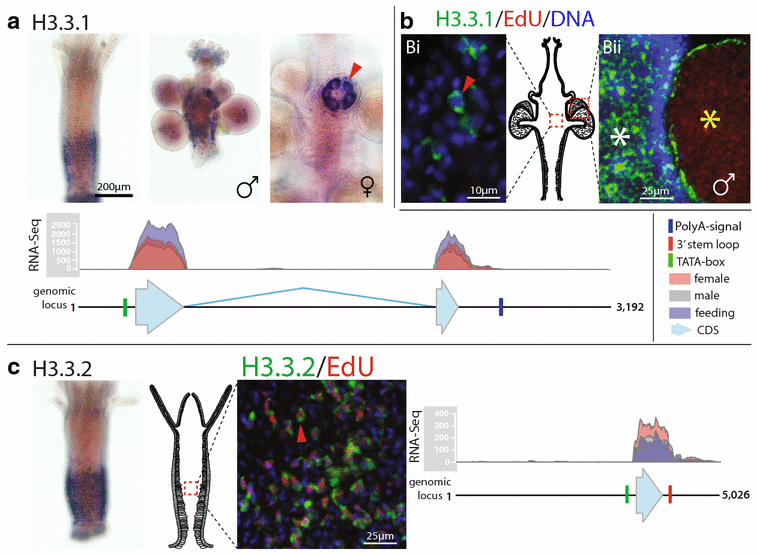
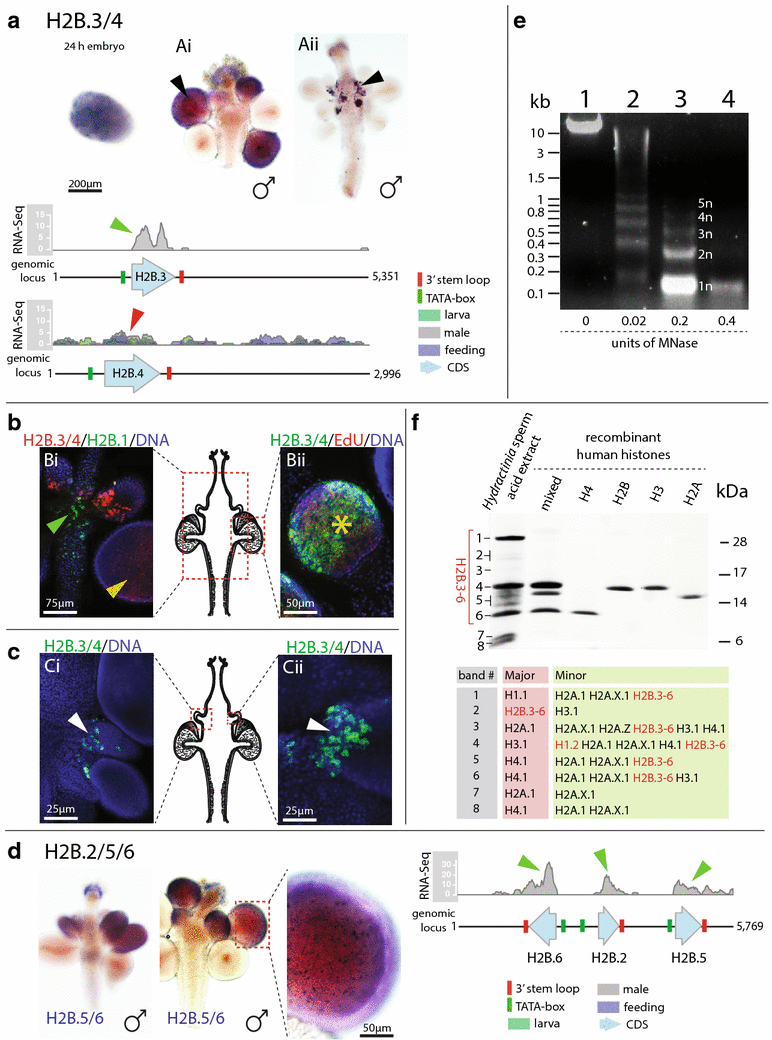
Results: We find that the Hydractinia genome encodes 19 histones and analyse their spatial expression patterns, genomic loci and replication-dependency. Alongside core and other replication-independent histone variants, we find several histone replication-dependent variants, including a rare replication-dependent H3.3, a female germ cell-specific H2A.X and an unusual set of five H2B variants, four of which are male germ cell-specific. We further confirm the absence of protamines in Hydractinia.
Conclusions: Since no protamines are found in hydroids, we suggest that the novel H2B variants are pivotal for sperm DNA packaging in this class of Cnidaria. This study adds to the limited number of full histone gene complements available in animals and sets a comprehensive framework for future studies on the role of histones and their post-translational modifications in cnidarian epigenetics. Finally, it provides insight into the evolution of spermatogenesis.
Keywords: Chromatin; Cnidaria; Histone; Histone variants; Sperm-specific histones.
Figures
Similar articles
-
Hydrozoan sperm-specific SPKK motif-containing histone H2B variants stabilise chromatin with limited compaction.Development. 2023 Jan 1;150(1):dev201058. doi: 10.1242/dev.201058. Epub 2023 Jan 12. Development. 2023. PMID: 36633190 Free PMC article.
-
Rapid divergence of histones in Hydrozoa (Cnidaria) and evolution of a novel histone involved in DNA damage response in hydra.Zoology (Jena). 2017 Aug;123:53-63. doi: 10.1016/j.zool.2017.06.005. Epub 2017 Jun 15. Zoology (Jena). 2017. PMID: 28720323
-
Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells.Epigenetics Chromatin. 2016 Jun 21;9:24. doi: 10.1186/s13072-016-0072-6. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27330565 Free PMC article.
-
Histone variants: key players of chromatin.Cell Tissue Res. 2014 Jun;356(3):457-66. doi: 10.1007/s00441-014-1862-4. Epub 2014 Apr 30. Cell Tissue Res. 2014. PMID: 24781148 Review.
-
The role of epigenetics in spermatogenesis.Turk J Urol. 2013 Sep;39(3):181-7. doi: 10.5152/tud.2013.037. Turk J Urol. 2013. PMID: 26328105 Free PMC article. Review.
Cited by
-
The Molecular Mechanisms Employed by the Parasite Myxobolus bejeranoi (Cnidaria: Myxozoa) from Invasion through Sporulation for Successful Proliferation in Its Fish Host.Int J Mol Sci. 2023 Aug 15;24(16):12824. doi: 10.3390/ijms241612824. Int J Mol Sci. 2023. PMID: 37629003 Free PMC article.
-
Acoel single-cell atlas reveals expression dynamics and heterogeneity of adult pluripotent stem cells.Nat Commun. 2023 May 5;14(1):2612. doi: 10.1038/s41467-023-38016-4. Nat Commun. 2023. PMID: 37147314 Free PMC article.
-
Parallels and contrasts between the cnidarian and bilaterian maternal-to-zygotic transition are revealed in Hydractinia embryos.bioRxiv [Preprint]. 2023 May 10:2023.05.09.540083. doi: 10.1101/2023.05.09.540083. bioRxiv. 2023. Update in: PLoS Genet. 2023 Jul 13;19(7):e1010845. doi: 10.1371/journal.pgen.1010845 PMID: 37214839 Free PMC article. Updated. Preprint.
-
The Biochemistry and Evolution of the Dinoflagellate Nucleus.Microorganisms. 2019 Aug 8;7(8):245. doi: 10.3390/microorganisms7080245. Microorganisms. 2019. PMID: 31398798 Free PMC article. Review.
-
Parallels and contrasts between the cnidarian and bilaterian maternal-to-zygotic transition are revealed in Hydractinia embryos.PLoS Genet. 2023 Jul 13;19(7):e1010845. doi: 10.1371/journal.pgen.1010845. eCollection 2023 Jul. PLoS Genet. 2023. PMID: 37440598 Free PMC article.
References
-
- Richmond TJ, Luger K, Mäder AW, Richmond RK, Sargent DF. Crystal structure of the nucleosome core particle at 2.8|[thinsp] [Aring]| resolution. Nature. Nature Publishing. Group. 1997;389:251–260. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

