Structural mechanism for the recognition and ubiquitination of a single nucleosome residue by Rad6-Bre1
- PMID: 27601672
- PMCID: PMC5035881
- DOI: 10.1073/pnas.1606863113
Structural mechanism for the recognition and ubiquitination of a single nucleosome residue by Rad6-Bre1
Abstract
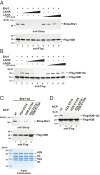
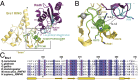
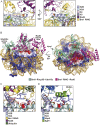
Cotranscriptional ubiquitination of histone H2B is key to gene regulation. The yeast E3 ubiquitin ligase Bre1 (human RNF20/40) pairs with the E2 ubiquitin conjugating enzyme Rad6 to monoubiquitinate H2B at Lys123. How this single lysine residue on the nucleosome core particle (NCP) is targeted by the Rad6-Bre1 machinery is unknown. Using chemical cross-linking and mass spectrometry, we identified the functional interfaces of Rad6, Bre1, and NCPs in a defined in vitro system. The Bre1 RING domain cross-links exclusively with distinct regions of histone H2B and H2A, indicating a spatial alignment of Bre1 with the NCP acidic patch. By docking onto the NCP surface in this distinct orientation, Bre1 positions the Rad6 active site directly over H2B Lys123. The Spt-Ada-Gcn5 acetyltransferase (SAGA) H2B deubiquitinase module competes with Bre1 for binding to the NCP acidic patch, indicating regulatory control. Our study reveals a mechanism that ensures site-specific NCP ubiquitination and fine-tuning of opposing enzymatic activities.
Keywords: Bre1–Rad6; RING E3 ligase; cross-linking mass spectrometry; nucleosome; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Monoubiquitination of histone H2B is intrinsic to the Bre1 RING domain-Rad6 interaction and augmented by a second Rad6-binding site on Bre1.J Biol Chem. 2015 Feb 27;290(9):5298-310. doi: 10.1074/jbc.M114.626788. Epub 2014 Dec 29. J Biol Chem. 2015. PMID: 25548288 Free PMC article.
-
Structural basis for the role of C-terminus acidic tail of Saccharomyces cerevisiae ubiquitin-conjugating enzyme (Rad6) in E3 ligase (Bre1) mediated recognition of histones.Int J Biol Macromol. 2024 Jan;254(Pt 2):127717. doi: 10.1016/j.ijbiomac.2023.127717. Epub 2023 Nov 2. Int J Biol Macromol. 2024. PMID: 37923031
-
Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.Mol Cell. 2023 Sep 7;83(17):3080-3094.e14. doi: 10.1016/j.molcel.2023.08.001. Epub 2023 Aug 25. Mol Cell. 2023. PMID: 37633270
-
The Bre1/Rad6 machinery: writing the central histone ubiquitin mark on H2B and beyond.Chromosome Res. 2020 Dec;28(3-4):247-258. doi: 10.1007/s10577-020-09640-3. Epub 2020 Sep 7. Chromosome Res. 2020. PMID: 32895784 Review.
-
H2B ubiquitination: Conserved molecular mechanism, diverse physiologic functions of the E3 ligase during meiosis.Nucleus. 2017 Sep 3;8(5):461-468. doi: 10.1080/19491034.2017.1330237. Epub 2017 Jun 19. Nucleus. 2017. PMID: 28628358 Free PMC article. Review.
Cited by
-
Structure of the human Bre1 complex bound to the nucleosome.Nat Commun. 2024 Mar 22;15(1):2580. doi: 10.1038/s41467-024-46910-8. Nat Commun. 2024. PMID: 38519511 Free PMC article.
-
Structural basis for the Rad6 activation by the Bre1 N-terminal domain.Elife. 2023 Mar 13;12:e84157. doi: 10.7554/eLife.84157. Elife. 2023. PMID: 36912886 Free PMC article.
-
Dynamic modules of the coactivator SAGA in eukaryotic transcription.Exp Mol Med. 2020 Jul;52(7):991-1003. doi: 10.1038/s12276-020-0463-4. Epub 2020 Jul 3. Exp Mol Med. 2020. PMID: 32616828 Free PMC article. Review.
-
The Paf1 Complex: A Keystone of Nuclear Regulation Operating at the Interface of Transcription and Chromatin.J Mol Biol. 2021 Jul 9;433(14):166979. doi: 10.1016/j.jmb.2021.166979. Epub 2021 Apr 1. J Mol Biol. 2021. PMID: 33811920 Free PMC article. Review.
-
Higher-order organization of biomolecular condensates.Open Biol. 2021 Jun;11(6):210137. doi: 10.1098/rsob.210137. Epub 2021 Jun 16. Open Biol. 2021. PMID: 34129784 Free PMC article. Review.
References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. - PubMed
-
- Wood A, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11(1):267–274. - PubMed
-
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287(5452):501–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

