Analysis of the machinery and intermediates of the 5hmC-mediated DNA demethylation pathway in aging on samples from the MARK-AGE Study
- PMID: 27587280
- PMCID: PMC5076444
- DOI: 10.18632/aging.101022
Analysis of the machinery and intermediates of the 5hmC-mediated DNA demethylation pathway in aging on samples from the MARK-AGE Study
Abstract
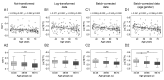
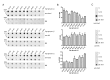
Gradual changes in the DNA methylation landscape occur throughout aging virtually in all human tissues. A widespread reduction of 5-methylcytosine (5mC), associated with highly reproducible site-specific hypermethylation, characterizes the genome in aging. Therefore, an equilibrium seems to exist between general and directional deregulating events concerning DNA methylation controllers, which may underpin the age-related epigenetic changes. In this context, 5mC-hydroxylases (TET enzymes) are new potential players. In fact, TETs catalyze the stepwise oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), driving the DNA demethylation process based on thymine DNA glycosylase (TDG)-mediated DNA repair pathway. The present paper reports the expression of DNA hydroxymethylation components, the levels of 5hmC and of its derivatives in peripheral blood mononuclear cells of age-stratified donors recruited in several European countries in the context of the EU Project 'MARK-AGE'. The results provide evidence for an age-related decline of TET1, TET3 and TDG gene expression along with a decrease of 5hmC and an accumulation of 5caC. These associations were independent of confounding variables, including recruitment center, gender and leukocyte composition. The observed impairment of 5hmC-mediated DNA demethylation pathway in blood cells may lead to aberrant transcriptional programs in the elderly.
Keywords: DNA hydroxymethylation; TDG; aging; genesTET.
Conflict of interest statement
All authors declare no conflict of interest.
Figures






Similar articles
-
Medulloblastoma and ependymoma cells display increased levels of 5-carboxylcytosine and elevated TET1 expression.Clin Epigenetics. 2017 Feb 13;9:18. doi: 10.1186/s13148-016-0306-2. eCollection 2017. Clin Epigenetics. 2017. PMID: 28228863 Free PMC article.
-
Structure and Function of TET Enzymes.Adv Exp Med Biol. 2016;945:275-302. doi: 10.1007/978-3-319-43624-1_12. Adv Exp Med Biol. 2016. PMID: 27826843 Review.
-
TET-mediated active DNA demethylation: mechanism, function and beyond.Nat Rev Genet. 2017 Sep;18(9):517-534. doi: 10.1038/nrg.2017.33. Epub 2017 May 30. Nat Rev Genet. 2017. PMID: 28555658 Review.
-
Ten-eleven translocation (Tet) and thymine DNA glycosylase (TDG), components of the demethylation pathway, are direct targets of miRNA-29a.Biochem Biophys Res Commun. 2013 Aug 2;437(3):368-73. doi: 10.1016/j.bbrc.2013.06.082. Epub 2013 Jun 29. Biochem Biophys Res Commun. 2013. PMID: 23820384 Free PMC article.
-
TET enzymes and 5hmC epigenetic mark: new key players in carcinogenesis and progression in gynecological cancers.Eur Rev Med Pharmacol Sci. 2024 Feb;28(3):1123-1134. doi: 10.26355/eurrev_202402_35349. Eur Rev Med Pharmacol Sci. 2024. PMID: 38375718 Review.
Cited by
-
Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials.Signal Transduct Target Ther. 2019 Dec 17;4:62. doi: 10.1038/s41392-019-0095-0. eCollection 2019. Signal Transduct Target Ther. 2019. PMID: 31871779 Free PMC article. Review.
-
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection: Triggering a Lethal Fight to Keep Control of the Ten-Eleven Translocase (TET)-Associated DNA Demethylation?Pathogens. 2020 Nov 30;9(12):1006. doi: 10.3390/pathogens9121006. Pathogens. 2020. PMID: 33266135 Free PMC article. Review.
-
Revisiting the genomic hypomethylation hypothesis of aging.Ann N Y Acad Sci. 2018 Apr;1418(1):69-79. doi: 10.1111/nyas.13533. Epub 2018 Jan 24. Ann N Y Acad Sci. 2018. PMID: 29363785 Free PMC article.
-
DNA Hydroxymethylation Levels Are Altered in Blood Cells From Down Syndrome Persons Enrolled in the MARK-AGE Project.J Gerontol A Biol Sci Med Sci. 2018 May 9;73(6):737-744. doi: 10.1093/gerona/glx198. J Gerontol A Biol Sci Med Sci. 2018. PMID: 29069286 Free PMC article.
-
Tet1 Deficiency Leads to Premature Reproductive Aging by Reducing Spermatogonia Stem Cells and Germ Cell Differentiation.iScience. 2020 Mar 27;23(3):100908. doi: 10.1016/j.isci.2020.100908. Epub 2020 Feb 13. iScience. 2020. PMID: 32114381 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

