Graft-Infiltrating Macrophages Adopt an M2 Phenotype and Are Inhibited by Purinergic Receptor P2X7 Antagonist in Chronic Rejection
- PMID: 27575724
- PMCID: PMC5552361
- DOI: 10.1111/ajt.13808
Graft-Infiltrating Macrophages Adopt an M2 Phenotype and Are Inhibited by Purinergic Receptor P2X7 Antagonist in Chronic Rejection
Abstract
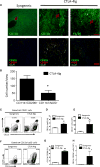
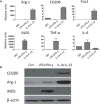
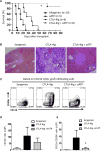
Macrophages exhibit diverse phenotypes and functions; they are also a major cell type infiltrating chronically rejected allografts. The exact phenotypes and roles of macrophages in chronic graft loss remain poorly defined. In the present study, we used a mouse heart transplant model to examine macrophages in chronic allograft rejection. We found that treatment of C57BL/6 mice with CTLA4 immunoglobulin fusion protein (CTLA4-Ig) prevented acute rejection of a Balb/c heart allograft but allowed chronic rejection to develop over time, characterized by prominent neointima formation in the graft. There was extensive macrophage infiltration in the chronically rejected allografts, and the graft-infiltrating macrophages expressed markers associated with M2 cells but not M1 cells. In an in vitro system in which macrophages were polarized into either M1 or M2 cells, we screened phenotypic differences between M1 and M2 cells and identified purinergic receptor P2X7 (P2x7r), an adenosine triphosphate (ATP)-gated ion channel protein that was preferentially expressed by M2 cells. We further showed that blocking the P2x7r using oxidized ATP (oATP) inhibited M2 induction in a dose-dependent fashion in vitro. Moreover, treatment of C57BL/6 recipients with the P2x7r antagonist oATP, in addition to CTLA4-Ig treatment, inhibited graft-infiltrating M2 cells, prevented transplant vasculopathy, and induced long-term heart allografts survival. These findings highlight the importance of the P2x7r-M2 axis in chronic rejection and establish P2x7r as a potential therapeutic target in suppression of chronic rejection.
Keywords: basic (laboratory) research/science; heart (allograft) function/dysfunction; heart transplantation/cardiology; macrophage/monocyte biology; rejection: chronic; vasculopathy.
© Copyright 2016 The American Society of Transplantation and the American Society of Transplant Surgeons.
Conflict of interest statement
The authors of this manuscript have no conflicts of interest to disclose as described by the
Figures



Comment in
-
Strategically Altering the Balance of Macrophage Subpopulations to Inhibit Chronic Rejection.Am J Transplant. 2016 Sep;16(9):2510-1. doi: 10.1111/ajt.13849. Epub 2016 Jun 9. Am J Transplant. 2016. PMID: 27136758 Free PMC article. No abstract available.
Similar articles
-
Macrophage subpopulations and their impact on chronic allograft rejection versus graft acceptance in a mouse heart transplant model.Am J Transplant. 2018 Mar;18(3):604-616. doi: 10.1111/ajt.14543. Epub 2017 Nov 23. Am J Transplant. 2018. PMID: 29044999 Free PMC article.
-
Inhibition of the purinergic pathway prolongs mouse lung allograft survival.Am J Respir Cell Mol Biol. 2014 Aug;51(2):300-10. doi: 10.1165/rcmb.2013-0362OC. Am J Respir Cell Mol Biol. 2014. PMID: 24661183
-
Long-term heart transplant survival by targeting the ionotropic purinergic receptor P2X7.Circulation. 2013 Jan 29;127(4):463-75. doi: 10.1161/CIRCULATIONAHA.112.123653. Epub 2012 Dec 18. Circulation. 2013. PMID: 23250993 Free PMC article.
-
M2 Macrophages Serve as Critical Executor of Innate Immunity in Chronic Allograft Rejection.Front Immunol. 2021 Mar 17;12:648539. doi: 10.3389/fimmu.2021.648539. eCollection 2021. Front Immunol. 2021. PMID: 33815407 Free PMC article. Review.
-
The P2X7 purinergic receptor: a potential therapeutic target for lung cancer.J Cancer Res Clin Oncol. 2020 Nov;146(11):2731-2741. doi: 10.1007/s00432-020-03379-4. Epub 2020 Sep 5. J Cancer Res Clin Oncol. 2020. PMID: 32892231 Review.
Cited by
-
MEK1/2-PKM2 Pathway Modulates the Immunometabolic Reprogramming of Proinflammatory Allograft-infiltrating Macrophages During Heart Transplant Rejection.Transplantation. 2024 May 1;108(5):1127-1141. doi: 10.1097/TP.0000000000004899. Epub 2024 Jan 19. Transplantation. 2024. PMID: 38238904 Free PMC article.
-
The Evolving Roles of Macrophages in Organ Transplantation.J Immunol Res. 2019 Apr 24;2019:5763430. doi: 10.1155/2019/5763430. eCollection 2019. J Immunol Res. 2019. PMID: 31179346 Free PMC article. Review.
-
Microbiota composition modulates inflammation and neointimal hyperplasia after arterial angioplasty.J Vasc Surg. 2020 Apr;71(4):1378-1389.e3. doi: 10.1016/j.jvs.2019.06.208. Epub 2020 Feb 5. J Vasc Surg. 2020. PMID: 32035769 Free PMC article.
-
Inflammatory macrophage-associated 3-gene signature predicts subclinical allograft injury and graft survival.JCI Insight. 2018 Jan 25;3(2):e95659. doi: 10.1172/jci.insight.95659. eCollection 2018 Jan 25. JCI Insight. 2018. PMID: 29367465 Free PMC article.
-
Recall features and allorecognition in innate immunity.Transpl Int. 2018 Jan;31(1):6-13. doi: 10.1111/tri.13073. Epub 2017 Oct 20. Transpl Int. 2018. PMID: 28926127 Free PMC article. Review.
References
-
- Stehlik J, Mehra MR, Sweet SC, et al. The International Society for Heart and Lung Transplantation Registries in the Era of Big Data With Global Reach. J Heart Lung Transplant. 2015;34:1225–1232. - PubMed
-
- Loupy A, Toquet C, Rouvier P, et al. Late Failing Heart Allografts: Pathology of Cardiac Allograft Vasculopathy and Association With Antibody-Mediated Rejection. Am J Transplant. 2016;16:111–120. - PubMed
-
- Tullius SG, Tilney NL. Both alloantigen-dependent and -independent factors influence chronic allograft rejection. Transplantation. 1995;59:313–318. - PubMed
-
- Meier-Kriesche HU, Schold JD, Kaplan B. Long-term renal allogrft survial: Have we made significant progress or is it time to rethink our analytic and therapeutic strategies? Am J Transplant. 2004;4:1289–1295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

