Single-Particle Refinement and Variability Analysis in EMAN2.1
- PMID: 27572727
- PMCID: PMC5101015
- DOI: 10.1016/bs.mie.2016.05.001
Single-Particle Refinement and Variability Analysis in EMAN2.1
Abstract
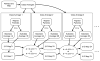
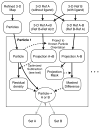
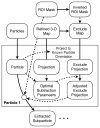
CryoEM single-particle reconstruction has been growing rapidly over the last 3 years largely due to the development of direct electron detectors, which have provided data with dramatic improvements in image quality. It is now possible in many cases to produce near-atomic resolution structures, and yet 2/3 of published structures remain at substantially lower resolutions. One important cause for this is compositional and conformational heterogeneity, which is both a resolution-limiting factor and presenting a unique opportunity to better relate structure to function. This manuscript discusses the canonical methods for high-resolution refinement in EMAN2.12, and then considers the wide range of available methods within this package for resolving structural variability, targeting both improved resolution and additional knowledge about particle dynamics.
Keywords: 3-D reconstruction; CryoEM; EMAN; Heterogeneity; Image processing; Motion; Single-particle analysis; Structural biology; Structural variability.
© 2016 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Testing the Validity of Single-Particle Maps at Low and High Resolution.Methods Enzymol. 2016;579:227-53. doi: 10.1016/bs.mie.2016.06.004. Epub 2016 Aug 8. Methods Enzymol. 2016. PMID: 27572729 Review.
-
Processing of Structurally Heterogeneous Cryo-EM Data in RELION.Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review.
-
Cryo-EM Structure Determination Using Segmented Helical Image Reconstruction.Methods Enzymol. 2016;579:307-28. doi: 10.1016/bs.mie.2016.05.034. Epub 2016 Jun 28. Methods Enzymol. 2016. PMID: 27572732 Review.
-
Tools for Model Building and Optimization into Near-Atomic Resolution Electron Cryo-Microscopy Density Maps.Methods Enzymol. 2016;579:255-76. doi: 10.1016/bs.mie.2016.06.003. Epub 2016 Aug 12. Methods Enzymol. 2016. PMID: 27572730 Free PMC article. Review.
-
Frealign: An Exploratory Tool for Single-Particle Cryo-EM.Methods Enzymol. 2016;579:191-226. doi: 10.1016/bs.mie.2016.04.013. Epub 2016 Jun 7. Methods Enzymol. 2016. PMID: 27572728 Free PMC article. Review.
Cited by
-
Three-Dimensional Structure of Cytochrome c Nitrite Reductase As Determined by Cryo-Electron Microscopy.Acta Naturae. 2018 Jul-Sep;10(3):48-56. Acta Naturae. 2018. PMID: 30397526 Free PMC article.
-
Multivalent Recruitment of Human Argonaute by GW182.Mol Cell. 2017 Aug 17;67(4):646-658.e3. doi: 10.1016/j.molcel.2017.07.007. Epub 2017 Aug 3. Mol Cell. 2017. PMID: 28781232 Free PMC article.
-
SIMPLE 3.0. Stream single-particle cryo-EM analysis in real time.J Struct Biol X. 2020 Nov 7;4:100040. doi: 10.1016/j.yjsbx.2020.100040. eCollection 2020. J Struct Biol X. 2020. PMID: 33294840 Free PMC article.
-
eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response.Science. 2019 May 3;364(6439):491-495. doi: 10.1126/science.aaw2922. Science. 2019. PMID: 31048491 Free PMC article.
-
New software tools in EMAN2 inspired by EMDatabank map challenge.J Struct Biol. 2018 Nov;204(2):283-290. doi: 10.1016/j.jsb.2018.09.002. Epub 2018 Sep 4. J Struct Biol. 2018. PMID: 30189321 Free PMC article.
References
-
- Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, et al. The crystal structure of the bacterial chaperonin groel at 2.8 Å. Nature. 1994;371(6498):578–86. - PubMed
-
- Brink J, Ludtke SJ, Kong Y, Wakil SJ, Ma J, Chiu W. Experimental verification of conformational variation of human fatty acid synthase as predicted by normal mode analysis. Structure (London, England : 1993) 2004;12(2):185–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

