Statistical Techniques Complement UML When Developing Domain Models of Complex Dynamical Biosystems
- PMID: 27571414
- PMCID: PMC5003378
- DOI: 10.1371/journal.pone.0160834
Statistical Techniques Complement UML When Developing Domain Models of Complex Dynamical Biosystems
Abstract
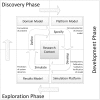
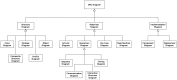
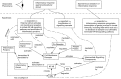
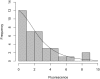
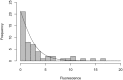
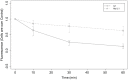
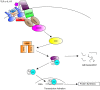
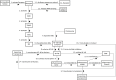
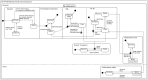
Computational modelling and simulation is increasingly being used to complement traditional wet-lab techniques when investigating the mechanistic behaviours of complex biological systems. In order to ensure computational models are fit for purpose, it is essential that the abstracted view of biology captured in the computational model, is clearly and unambiguously defined within a conceptual model of the biological domain (a domain model), that acts to accurately represent the biological system and to document the functional requirements for the resultant computational model. We present a domain model of the IL-1 stimulated NF-κB signalling pathway, which unambiguously defines the spatial, temporal and stochastic requirements for our future computational model. Through the development of this model, we observe that, in isolation, UML is not sufficient for the purpose of creating a domain model, and that a number of descriptive and multivariate statistical techniques provide complementary perspectives, in particular when modelling the heterogeneity of dynamics at the single-cell level. We believe this approach of using UML to define the structure and interactions within a complex system, along with statistics to define the stochastic and dynamic nature of complex systems, is crucial for ensuring that conceptual models of complex dynamical biosystems, which are developed using UML, are fit for purpose, and unambiguously define the functional requirements for the resultant computational model.
Conflict of interest statement
The authors of this manuscript have read the journal’s policy and have the following competing interests: JT is Director of SimOmics Ltd. This does not alter the authors’ adherence to PLOS ONE policies on sharing data and materials.
Figures


Similar articles
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Using the Unified Modelling Language (UML) to guide the systemic description of biological processes and systems.Biosystems. 2004 Jul;75(1-3):3-14. doi: 10.1016/j.biosystems.2004.03.009. Biosystems. 2004. PMID: 15245800
-
Modelling biological behaviours with the unified modelling language: an immunological case study and critique.J R Soc Interface. 2014 Oct 6;11(99):20140704. doi: 10.1098/rsif.2014.0704. J R Soc Interface. 2014. PMID: 25142524 Free PMC article.
-
Models of signalling networks - what cell biologists can gain from them and give to them.J Cell Sci. 2013 May 1;126(Pt 9):1913-21. doi: 10.1242/jcs.112045. J Cell Sci. 2013. PMID: 23720376 Free PMC article. Review.
-
Engineering simulations for cancer systems biology.Curr Drug Targets. 2012 Nov;13(12):1560-74. doi: 10.2174/138945012803530071. Curr Drug Targets. 2012. PMID: 22974398 Review.
Cited by
-
Designing and evaluating the children's developmental motor disorders system: an experience from a developing country.BMC Med Inform Decis Mak. 2023 Jul 17;23(1):123. doi: 10.1186/s12911-023-02223-2. BMC Med Inform Decis Mak. 2023. PMID: 37455319 Free PMC article.
-
Rethinking gamete donor care: A satisfaction survey of egg and sperm donors in the UK.PLoS One. 2018 Jul 23;13(7):e0199971. doi: 10.1371/journal.pone.0199971. eCollection 2018. PLoS One. 2018. PMID: 30036373 Free PMC article.
References
-
- Andrews PS, Polack FAC, Sampson AT, Stepney S, Timmis J (2010) The CoSMoS process, version 0.1: A process for the modelling and simulation of complex systems. Tech. Rep. YCS-2010-453, University of York
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

