The BCR-ABL/NF-κB signal transduction network: a long lasting relationship in Philadelphia positive Leukemias
- PMID: 27563822
- PMCID: PMC5323234
- DOI: 10.18632/oncotarget.11507
The BCR-ABL/NF-κB signal transduction network: a long lasting relationship in Philadelphia positive Leukemias
Abstract
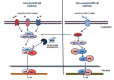
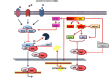
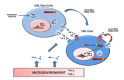
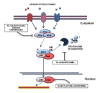
The Nuclear Factor-kappa B (NF-κB) family of transcription factors plays a key role in cancer pathogenesis due to the ability to promote cellular proliferation and survival, to induce resistance to chemotherapy and to mediate invasion and metastasis. NF-κB is recruited through different mechanisms involving either canonical (RelA/p50) or non-canonical pathways (RelB/p50 or RelB/p52), which transduce the signals originated from growth-factors, cytokines, oncogenic stress and DNA damage, bacterial and viral products or other stimuli. The pharmacological inhibition of the NF-κB pathway has clearly been associated with significant clinical activity in different cancers. Almost 20 years ago, NF-κB was described as an essential modulator of BCR-ABL signaling in Chronic Myeloid Leukemia and Philadelphia-positive Acute Lymphoblastic Leukemia. This review summarizes the role of NF-κB in BCR-ABL-mediated leukemogenesis and provides new insights on the long lasting BCR-ABL/NF-κB connection.
Keywords: BCR-ABL; CML; IκB-α; NF-κB; NFKBIA.
Conflict of interest statement
The authors declare no competing financial interests to disclose.
Figures
Similar articles
-
IKK-dependent activation of NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1.Blood. 2014 Apr 10;123(15):2401-11. doi: 10.1182/blood-2014-01-547943. Epub 2014 Jan 24. Blood. 2014. PMID: 24464015 Free PMC article.
-
A requirement for NF-kappaB activation in Bcr-Abl-mediated transformation.Genes Dev. 1998 Apr 1;12(7):968-81. doi: 10.1101/gad.12.7.968. Genes Dev. 1998. PMID: 9531535 Free PMC article.
-
Constitutive NF-kappab/Rel activation in philadelphia chromosome positive (Ph+) acute lymphoblastic leukemia (ALL).Leuk Lymphoma. 2004 Jun;45(6):1181-4. doi: 10.1080/10428190310001657326. Leuk Lymphoma. 2004. PMID: 15359998
-
Regulation of mammalian target of rapamycin and mitogen activated protein kinase pathways by BCR-ABL.Leuk Lymphoma. 2011 Feb;52 Suppl 1:45-53. doi: 10.3109/10428194.2010.546919. Leuk Lymphoma. 2011. PMID: 21299459 Review.
-
The Philadelphia chromosome in leukemogenesis.Chin J Cancer. 2016 May 27;35:48. doi: 10.1186/s40880-016-0108-0. Chin J Cancer. 2016. PMID: 27233483 Free PMC article. Review.
Cited by
-
miR-181d/RBP2/NF-κB p65 Feedback Regulation Promotes Chronic Myeloid Leukemia Blast Crisis.Front Oncol. 2021 Mar 25;11:654411. doi: 10.3389/fonc.2021.654411. eCollection 2021. Front Oncol. 2021. PMID: 33842368 Free PMC article.
-
IκB-α: At the crossroad between oncogenic and tumor-suppressive signals.Oncol Lett. 2017 Feb;13(2):531-534. doi: 10.3892/ol.2016.5465. Epub 2016 Dec 6. Oncol Lett. 2017. PMID: 28356925 Free PMC article.
-
Irf8 regulates the progression of myeloproliferative neoplasm-like syndrome via Mertk signaling in zebrafish.Leukemia. 2018 Jan;32(1):149-158. doi: 10.1038/leu.2017.189. Epub 2017 Jun 19. Leukemia. 2018. PMID: 28626217
-
Targeting the MAPK/ERK and PI3K/AKT Signaling Pathways Affects NRF2, Trx and GSH Antioxidant Systems in Leukemia Cells.Antioxidants (Basel). 2020 Jul 17;9(7):633. doi: 10.3390/antiox9070633. Antioxidants (Basel). 2020. PMID: 32709140 Free PMC article.
-
Adaptor protein Abelson interactor 1 in homeostasis and disease.Cell Commun Signal. 2024 Oct 1;22(1):468. doi: 10.1186/s12964-024-01738-z. Cell Commun Signal. 2024. PMID: 39354505 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

