LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea
- PMID: 27559308
- PMCID: PMC4988241
- DOI: 10.3389/fncel.2016.00186
LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea
Abstract
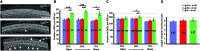
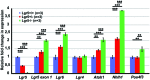
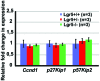
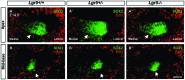
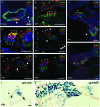
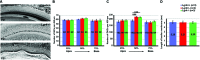
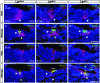
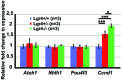
In the developing cochlea, Wnt/β-catenin signaling positively regulates the proliferation of precursors and promotes the formation of hair cells by up-regulating Atoh1 expression. Not much, however, is known about the regulation of Wnt/β-catenin activity in the cochlea. In multiple tissues, the activity of Wnt/β-catenin signaling is modulated by an interaction between LGR receptors and their ligands from the R-spondin family. The deficiency in Lgr4 and Lgr5 genes leads to developmental malformations and lethality. Using the Lgr5 knock-in mouse line we show that loss of LGR5 function increases Wnt/β-catenin activity in the embryonic cochlea, resulting in a mild overproduction of inner and outer hair cells (OHC). Supernumerary hair cells are likely formed due to an up-regulation of the "pro-hair cell" transcription factors Atoh1, Nhlh1, and Pou4f3. Using a hypomorphic Lgr4 mouse model we showed a mild overproduction of OHCs in the heterozygous and homozygous Lgr4 mice. The loss of LGR4 function prolonged the proliferation in the mid-basal turn of E13 cochleae, causing an increase in the number of SOX2-positive precursor cells within the pro-sensory domain. The premature differentiation of hair cells progressed in a medial to lateral gradient in Lgr4 deficient embryos. No significant up-regulation of Atoh1 was observed following Lgr4 deletion. Altogether, our findings suggest that LGR4 and LGR5 play an important role in the regulation of hair cell differentiation in the embryonic cochlea.
Keywords: LGR4; LGR5; Wnt signaling; cochlea; development; hair cells.
Figures
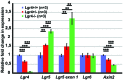
Similar articles
-
Functional roles of Lgr4 and Lgr5 in embryonic gut, kidney and skin development in mice.Dev Biol. 2014 Jun 15;390(2):181-90. doi: 10.1016/j.ydbio.2014.03.009. Epub 2014 Mar 26. Dev Biol. 2014. PMID: 24680895
-
Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea.Proc Natl Acad Sci U S A. 2012 May 22;109(21):8167-72. doi: 10.1073/pnas.1202774109. Epub 2012 May 4. Proc Natl Acad Sci U S A. 2012. PMID: 22562792 Free PMC article.
-
Extensive Supporting Cell Proliferation and Mitotic Hair Cell Generation by In Vivo Genetic Reprogramming in the Neonatal Mouse Cochlea.J Neurosci. 2016 Aug 17;36(33):8734-45. doi: 10.1523/JNEUROSCI.0060-16.2016. J Neurosci. 2016. PMID: 27535918 Free PMC article.
-
Atoh1 regulation in the cochlea: more than just transcription.J Zhejiang Univ Sci B. 2019 Feb.;20(2):146-155. doi: 10.1631/jzus.B1600438. Epub 2017 Jul 13. J Zhejiang Univ Sci B. 2019. PMID: 29770645 Free PMC article. Review.
-
Role of Wnt and Notch signaling in regulating hair cell regeneration in the cochlea.Front Med. 2016 Sep;10(3):237-49. doi: 10.1007/s11684-016-0464-9. Epub 2016 Sep 7. Front Med. 2016. PMID: 27527363 Review.
Cited by
-
G protein-coupled receptors in cochlea: Potential therapeutic targets for hearing loss.Front Mol Neurosci. 2022 Oct 12;15:1028125. doi: 10.3389/fnmol.2022.1028125. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36311029 Free PMC article. Review.
-
Lgr4 Regulates Oviductal Epithelial Secretion Through the WNT Signaling Pathway.Front Cell Dev Biol. 2021 Sep 24;9:666303. doi: 10.3389/fcell.2021.666303. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34631693 Free PMC article.
-
Open chromatin dynamics in prosensory cells of the embryonic mouse cochlea.Sci Rep. 2019 Jun 21;9(1):9060. doi: 10.1038/s41598-019-45515-2. Sci Rep. 2019. PMID: 31227770 Free PMC article.
-
LGR5-Positive Supporting Cells Survive Ototoxic Trauma in the Adult Mouse Cochlea.Front Mol Neurosci. 2021 Oct 5;14:729625. doi: 10.3389/fnmol.2021.729625. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34675775 Free PMC article.
-
Hedgehog Signaling Promotes the Proliferation and Subsequent Hair Cell Formation of Progenitor Cells in the Neonatal Mouse Cochlea.Front Mol Neurosci. 2017 Dec 21;10:426. doi: 10.3389/fnmol.2017.00426. eCollection 2017. Front Mol Neurosci. 2017. PMID: 29311816 Free PMC article.
References
-
- Adam J., Myat A., Le Roux I., Eddison M., Henrique D., Ish-Horowicz D., et al. (1998). Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development 125 4645–4654. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

