YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex
- PMID: 27558897
- PMCID: PMC5007331
- DOI: 10.1038/ncomms12626
YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex
Abstract
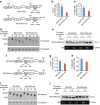
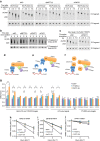
Methylation at the N6 position of adenosine (m(6)A) is the most abundant RNA modification within protein-coding and long noncoding RNAs in eukaryotes and is a reversible process with important biological functions. YT521-B homology domain family (YTHDF) proteins are the readers of m(6)A, the binding of which results in the alteration of the translation efficiency and stability of m(6)A-containing RNAs. However, the mechanism by which YTHDF proteins cause the degradation of m(6)A-containing RNAs is poorly understood. Here we report that m(6)A-containing RNAs exhibit accelerated deadenylation that is mediated by the CCR4-NOT deadenylase complex. We further show that YTHDF2 recruits the CCR4-NOT complex through a direct interaction between the YTHDF2 N-terminal region and the SH domain of the CNOT1 subunit, and that this recruitment is essential for the deadenylation of m(6)A-containing RNAs by CAF1 and CCR4. Therefore, we have uncovered the mechanism of YTHDF2-mediated degradation of m(6)A-containing RNAs in mammalian cells.
Figures



Similar articles
-
Molecular Mechanisms Driving mRNA Degradation by m6A Modification.Trends Genet. 2020 Mar;36(3):177-188. doi: 10.1016/j.tig.2019.12.007. Epub 2020 Jan 18. Trends Genet. 2020. PMID: 31964509 Review.
-
YTHDF2/3 Are Required for Somatic Reprogramming through Different RNA Deadenylation Pathways.Cell Rep. 2020 Sep 8;32(10):108120. doi: 10.1016/j.celrep.2020.108120. Cell Rep. 2020. PMID: 32905781
-
PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay.Mol Cell. 2018 Jun 21;70(6):1081-1088.e5. doi: 10.1016/j.molcel.2018.05.009. Mol Cell. 2018. PMID: 29932901
-
Tryptophan-Mediated Interactions between Tristetraprolin and the CNOT9 Subunit Are Required for CCR4-NOT Deadenylase Complex Recruitment.J Mol Biol. 2018 Mar 2;430(5):722-736. doi: 10.1016/j.jmb.2017.12.018. Epub 2017 Dec 29. J Mol Biol. 2018. PMID: 29291391
-
The Not3/5 subunit of the Ccr4-Not complex: a central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells.Cell Signal. 2013 Apr;25(4):743-51. doi: 10.1016/j.cellsig.2012.12.018. Epub 2012 Dec 29. Cell Signal. 2013. PMID: 23280189 Review.
Cited by
-
Transcriptome-wide N6-methyladenosine (m6A) methylation in soybean under Meloidogyne incognita infection.aBIOTECH. 2022 Aug 18;3(3):197-211. doi: 10.1007/s42994-022-00077-2. eCollection 2022 Sep. aBIOTECH. 2022. PMID: 36313932 Free PMC article.
-
RNA m6A Methyltransferase Mettl3 Regulates Spatial Neural Patterning in Xenopus laevis.Mol Cell Biol. 2021 Jul 23;41(8):e0010421. doi: 10.1128/MCB.00104-21. Epub 2021 Jul 23. Mol Cell Biol. 2021. PMID: 33972392 Free PMC article.
-
Mettl3-mediated m6A modification of Fgf16 restricts cardiomyocyte proliferation during heart regeneration.Elife. 2022 Nov 18;11:e77014. doi: 10.7554/eLife.77014. Elife. 2022. PMID: 36399125 Free PMC article.
-
N6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling.J Biol Chem. 2020 Sep 11;295(37):13123-13133. doi: 10.1074/jbc.RA120.014260. Epub 2020 Jul 27. J Biol Chem. 2020. PMID: 32719095 Free PMC article.
-
Dynamics of m6A RNA Methylome on the Hallmarks of Hepatocellular Carcinoma.Front Cell Dev Biol. 2021 Apr 1;9:642443. doi: 10.3389/fcell.2021.642443. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869193 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

