RNA-sequencing of a mouse-model of spinal muscular atrophy reveals tissue-wide changes in splicing of U12-dependent introns
- PMID: 27557711
- PMCID: PMC5224493
- DOI: 10.1093/nar/gkw731
RNA-sequencing of a mouse-model of spinal muscular atrophy reveals tissue-wide changes in splicing of U12-dependent introns
Abstract
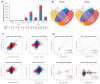
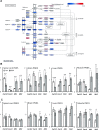
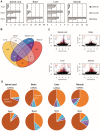
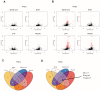
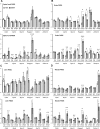
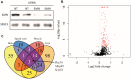
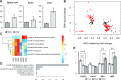
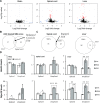
Spinal Muscular Atrophy (SMA) is a neuromuscular disorder caused by insufficient levels of the Survival of Motor Neuron (SMN) protein. SMN is expressed ubiquitously and functions in RNA processing pathways that include trafficking of mRNA and assembly of snRNP complexes. Importantly, SMA severity is correlated with decreased snRNP assembly activity. In particular, the minor spliceosomal snRNPs are affected, and some U12-dependent introns have been reported to be aberrantly spliced in patient cells and animal models. SMA is characterized by loss of motor neurons, but the underlying mechanism is largely unknown. It is likely that aberrant splicing of genes expressed in motor neurons is involved in SMA pathogenesis, but increasing evidence indicates that pathologies also exist in other tissues. We present here a comprehensive RNA-seq study that covers multiple tissues in an SMA mouse model. We show elevated U12-intron retention in all examined tissues from SMA mice, and that U12-dependent intron retention is induced upon siRNA knock-down of SMN in HeLa cells. Furthermore, we show that retention of U12-dependent introns is mitigated by ASO treatment of SMA mice and that many transcriptional changes are reversed. Finally, we report on missplicing of several Ca2+ channel genes that may explain disrupted Ca2+ homeostasis in SMA and activation of Cdk5.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage.Proc Natl Acad Sci U S A. 2017 Mar 21;114(12):E2347-E2356. doi: 10.1073/pnas.1613181114. Epub 2017 Mar 7. Proc Natl Acad Sci U S A. 2017. PMID: 28270613 Free PMC article.
-
Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model.Nature. 2011 Oct 5;478(7367):123-6. doi: 10.1038/nature10485. Nature. 2011. PMID: 21979052 Free PMC article.
-
Altered mRNA Splicing in SMN-Depleted Motor Neuron-Like Cells.PLoS One. 2016 Oct 13;11(10):e0163954. doi: 10.1371/journal.pone.0163954. eCollection 2016. PLoS One. 2016. PMID: 27736905 Free PMC article.
-
Spinal muscular atrophy: antisense oligonucleotide therapy opens the door to an integrated therapeutic landscape.Hum Mol Genet. 2017 Oct 1;26(R2):R151-R159. doi: 10.1093/hmg/ddx215. Hum Mol Genet. 2017. PMID: 28977438 Review.
-
SMN in spinal muscular atrophy and snRNP biogenesis.Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):546-64. doi: 10.1002/wrna.76. Epub 2011 Feb 17. Wiley Interdiscip Rev RNA. 2011. PMID: 21957043 Review.
Cited by
-
Splicing efficiency of minor introns in a mouse model of SMA predominantly depends on their branchpoint sequence and can involve the contribution of major spliceosome components.RNA. 2022 Mar;28(3):303-319. doi: 10.1261/rna.078329.120. Epub 2021 Dec 10. RNA. 2022. PMID: 34893560 Free PMC article.
-
Small-molecule flunarizine increases SMN protein in nuclear Cajal bodies and motor function in a mouse model of spinal muscular atrophy.Sci Rep. 2018 Feb 1;8(1):2075. doi: 10.1038/s41598-018-20219-1. Sci Rep. 2018. PMID: 29391529 Free PMC article.
-
Downregulation of Survivin contributes to cell-cycle arrest during postnatal cardiac development in a severe spinal muscular atrophy mouse model.Hum Mol Genet. 2018 Feb 1;27(3):486-498. doi: 10.1093/hmg/ddx418. Hum Mol Genet. 2018. PMID: 29220503 Free PMC article.
-
Increased systemic HSP70B levels in spinal muscular atrophy infants.Ann Clin Transl Neurol. 2021 Jul;8(7):1495-1501. doi: 10.1002/acn3.51377. Epub 2021 May 15. Ann Clin Transl Neurol. 2021. PMID: 33991176 Free PMC article.
-
Therapy development for spinal muscular atrophy: perspectives for muscular dystrophies and neurodegenerative disorders.Neurol Res Pract. 2022 Jan 4;4(1):2. doi: 10.1186/s42466-021-00162-9. Neurol Res Pract. 2022. PMID: 34983696 Free PMC article. Review.
References
-
- Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. - PubMed
-
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

