Protein arginine methylation/demethylation and cancer
- PMID: 27556302
- PMCID: PMC5341895
- DOI: 10.18632/oncotarget.11376
Protein arginine methylation/demethylation and cancer
Abstract
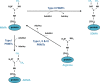
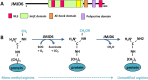
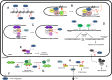
Protein arginine methylation is a common post-translational modification involved in numerous cellular processes including transcription, DNA repair, mRNA splicing and signal transduction. Currently, there are nine known members of the protein arginine methyltransferase (PRMT) family, but only one arginine demethylase has been identified, namely the Jumonji domain-containing 6 (JMJD6). Although its demethylase activity was initially challenged, its dual activity as an arginine demethylase and a lysine hydroxylase is now recognized. Interestingly, a growing number of substrates for arginine methylation and demethylation play key roles in tumorigenesis. Though alterations in the sequence of these enzymes have not been identified in cancer, their overexpression is associated with various cancers, suggesting that they could constitute targets for therapeutic strategies. In this review, we present the recent knowledge of the involvement of PRMTs and JMJD6 in tumorigenesis.
Keywords: JMJD6; PRMT; cancer; demethylation; methylation.
Conflict of interest statement
The authors declare no conflict of interest
Figures
Similar articles
-
Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1.J Biol Chem. 2017 Nov 17;292(46):18886-18896. doi: 10.1074/jbc.M117.800706. Epub 2017 Sep 27. J Biol Chem. 2017. PMID: 28972166 Free PMC article.
-
Elucidating the role of PRMTs in prostate cancer using open access databases and a patient cohort dataset.Histol Histopathol. 2023 Mar;38(3):287-302. doi: 10.14670/HH-18-513. Epub 2022 Sep 9. Histol Histopathol. 2023. PMID: 36082942
-
Insights into Jumonji C-domain containing protein 6 (JMJD6): a multifactorial role in foot-and-mouth disease virus replication in cells.Virus Genes. 2017 Jun;53(3):340-351. doi: 10.1007/s11262-017-1449-8. Epub 2017 Mar 31. Virus Genes. 2017. PMID: 28364140 Review.
-
JMJD6 regulates ERα methylation on arginine.PLoS One. 2014 Feb 3;9(2):e87982. doi: 10.1371/journal.pone.0087982. eCollection 2014. PLoS One. 2014. PMID: 24498420 Free PMC article.
-
Jumonji domain-containing protein 6 protein and its role in cancer.Cell Prolif. 2020 Feb;53(2):e12747. doi: 10.1111/cpr.12747. Epub 2020 Jan 21. Cell Prolif. 2020. PMID: 31961032 Free PMC article. Review.
Cited by
-
Epigenetics in ovarian cancer: premise, properties, and perspectives.Mol Cancer. 2018 Jul 31;17(1):109. doi: 10.1186/s12943-018-0855-4. Mol Cancer. 2018. PMID: 30064416 Free PMC article. Review.
-
Protein Arginine Methyltransferase (PRMT) Inhibitors-AMI-1 and SAH Are Effective in Attenuating Rhabdomyosarcoma Growth and Proliferation in Cell Cultures.Int J Mol Sci. 2021 Jul 27;22(15):8023. doi: 10.3390/ijms22158023. Int J Mol Sci. 2021. PMID: 34360791 Free PMC article.
-
Guanidine N-methylation by BlsL Is Dependent on Acylation of Beta-amine Arginine in the Biosynthesis of Blasticidin S.Front Microbiol. 2017 Aug 22;8:1565. doi: 10.3389/fmicb.2017.01565. eCollection 2017. Front Microbiol. 2017. PMID: 28878744 Free PMC article.
-
Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1.J Biol Chem. 2017 Nov 17;292(46):18886-18896. doi: 10.1074/jbc.M117.800706. Epub 2017 Sep 27. J Biol Chem. 2017. PMID: 28972166 Free PMC article.
-
Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story.Biomolecules. 2017 Jun 1;7(2):41. doi: 10.3390/biom7020041. Biomolecules. 2017. PMID: 28587176 Free PMC article. Review.
References
-
- Ambler RP, Rees MW. Epsilon-N-Methyl-lysine in bacterial flagellar protein. Nature. 1959;184:56–57. - PubMed
-
- Murray K. The occurrence of epsilon-n-methyl lysine in histones. Biochemistry. 1964;3:10–15. - PubMed
-
- Baldwin GS, Carnegie PR. Specific enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science. 1971;171(3971):579–581. - PubMed
-
- Kakimoto Y, Akazawa S. Isolation and identification of N-G,N-G- and N-G,N’-G-dimethyl-arginine, N-epsilon-mono-, di-, and trimethyllysine, and glucosylgalactosyl- and galactosyl-delta-hydroxylysine from human urine. J Biol Chem. 1970;245(21):5751–5758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

