Protein-DNA and ion-DNA interactions revealed through contrast variation SAXS
- PMID: 27551324
- PMCID: PMC4991782
- DOI: 10.1007/s12551-016-0196-8
Protein-DNA and ion-DNA interactions revealed through contrast variation SAXS
Abstract
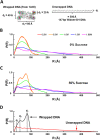
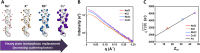
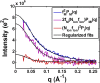
Understanding how DNA carries out its biological roles requires knowledge of its interactions with biological partners. Since DNA is a polyanionic polymer, electrostatic interactions contribute significantly. These interactions are mediated by positively charged protein residues or charge compensating cations. Direct detection of these partners and/or their effect on DNA conformation poses challenges, especially for monitoring conformational dynamics in real time. Small-angle x-ray scattering (SAXS) is uniquely sensitive to both the conformation and local environment (i.e. protein partner and associated ions) of the DNA. The primary challenge of studying multi-component systems with SAXS lies in resolving how each component contributes to the measured scattering. Here, we review two contrast variation (CV) strategies that enable targeted studies of the structures of DNA or its associated partners. First, solution contrast variation enables measurement of DNA conformation within a protein-DNA complex by masking out the protein contribution to the scattering profile. We review a specific example, in which the real-time unwrapping of DNA from a nucleosome core particle is measured during salt-induced disassembly. The second method, heavy atom isomorphous replacement, reports the spatial distribution of the cation cloud around duplex DNA by exploiting changes in the scattering strength of cations with varying atomic numbers. We demonstrate the application of this approach to provide the spatial distribution of monovalent cations (Na+, K+, Rb+, Cs+) around a standard 25-base pair DNA. The CV strategies presented here are valuable tools for understanding DNA interactions with its biological partners.
Keywords: Contrast variation; DNA; Heavy atom isomorphous replacement; Ions; NCP; SAXS.
Conflict of interest statement
Conflict of interest
Joshua M. Tokuda declares that he has no conflict of interest.
Suzette A. Pabit declares that she has no conflict of interest.
Lois Pollack declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures



Similar articles
-
Determining the Locations of Ions and Water around DNA from X-Ray Scattering Measurements.Biophys J. 2015 Jun 16;108(12):2886-95. doi: 10.1016/j.bpj.2015.05.006. Biophys J. 2015. PMID: 26083928 Free PMC article.
-
Counterion distribution surrounding spherical nucleic acid-Au nanoparticle conjugates probed by small-angle x-ray scattering.ACS Nano. 2013 Dec 23;7(12):11301-9. doi: 10.1021/nn405109z. Epub 2013 Nov 23. ACS Nano. 2013. PMID: 24251367
-
An advanced coarse-grained nucleosome core particle model for computer simulations of nucleosome-nucleosome interactions under varying ionic conditions.PLoS One. 2013;8(2):e54228. doi: 10.1371/journal.pone.0054228. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418426 Free PMC article.
-
SAXS studies of ion-nucleic acid interactions.Annu Rev Biophys. 2011;40:225-42. doi: 10.1146/annurev-biophys-042910-155349. Annu Rev Biophys. 2011. PMID: 21332357 Review.
-
SAXS studies of RNA: structures, dynamics, and interactions with partners.Wiley Interdiscip Rev RNA. 2016 Jul;7(4):512-26. doi: 10.1002/wrna.1349. Epub 2016 Apr 12. Wiley Interdiscip Rev RNA. 2016. PMID: 27071649 Free PMC article. Review.
Cited by
-
Revealing the distinct folding phases of an RNA three-helix junction.Nucleic Acids Res. 2018 Aug 21;46(14):7354-7365. doi: 10.1093/nar/gky363. Nucleic Acids Res. 2018. PMID: 29762712 Free PMC article.
-
Local DNA Sequence Controls Asymmetry of DNA Unwrapping from Nucleosome Core Particles.Biophys J. 2018 Sep 4;115(5):773-781. doi: 10.1016/j.bpj.2018.07.009. Epub 2018 Jul 31. Biophys J. 2018. PMID: 30072033 Free PMC article.
-
Contrast variation SAXS: Sample preparation protocols, experimental procedures, and data analysis.Methods Enzymol. 2022;677:41-83. doi: 10.1016/bs.mie.2022.08.007. Epub 2022 Sep 22. Methods Enzymol. 2022. PMID: 36410957 Free PMC article.
-
Nucleosome structure and dynamics are coming of age.Nat Struct Mol Biol. 2019 Jan;26(1):3-13. doi: 10.1038/s41594-018-0166-x. Epub 2018 Dec 10. Nat Struct Mol Biol. 2019. PMID: 30532059 Free PMC article. Review.
-
Medical contrast media as possible tools for SAXS contrast variation.IUCrJ. 2019 May 29;6(Pt 4):521-525. doi: 10.1107/S2052252519005943. eCollection 2019 Jul 1. IUCrJ. 2019. PMID: 31316796 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

