Lincosamides, Streptogramins, Phenicols, and Pleuromutilins: Mode of Action and Mechanisms of Resistance
- PMID: 27549310
- PMCID: PMC5088508
- DOI: 10.1101/cshperspect.a027037
Lincosamides, Streptogramins, Phenicols, and Pleuromutilins: Mode of Action and Mechanisms of Resistance
Abstract
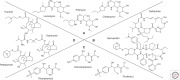
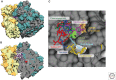
Lincosamides, streptogramins, phenicols, and pleuromutilins (LSPPs) represent four structurally different classes of antimicrobial agents that inhibit bacterial protein synthesis by binding to particular sites on the 50S ribosomal subunit of the ribosomes. Members of all four classes are used for different purposes in human and veterinary medicine in various countries worldwide. Bacteria have developed ways and means to escape the inhibitory effects of LSPP antimicrobial agents by enzymatic inactivation, active export, or modification of the target sites of the agents. This review provides a comprehensive overview of the mode of action of LSPP antimicrobial agents as well as of the mutations and resistance genes known to confer resistance to these agents in various bacteria of human and animal origin.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures



Similar articles
-
Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70.Antimicrob Agents Chemother. 2011 Apr;55(4):1470-4. doi: 10.1128/AAC.01068-10. Epub 2011 Jan 18. Antimicrob Agents Chemother. 2011. PMID: 21245447 Free PMC article.
-
Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.Nat Commun. 2021 Jun 11;12(1):3577. doi: 10.1038/s41467-021-23753-1. Nat Commun. 2021. PMID: 34117249 Free PMC article.
-
Detailed mutational analysis of Vga(A) interdomain linker: implication for antibiotic resistance specificity and mechanism.Antimicrob Agents Chemother. 2015 Feb;59(2):1360-4. doi: 10.1128/AAC.04468-14. Epub 2014 Dec 15. Antimicrob Agents Chemother. 2015. PMID: 25512423 Free PMC article.
-
Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage - a review.Ann Agric Environ Med. 2017 Jun 12;24(2):338-344. doi: 10.26444/aaem/74718. Epub 2017 Jun 12. Ann Agric Environ Med. 2017. PMID: 28664720 Review.
-
Pleuromutilins: Potent Drugs for Resistant Bugs-Mode of Action and Resistance.Cold Spring Harb Perspect Med. 2017 Jan 3;7(1):a027110. doi: 10.1101/cshperspect.a027110. Cold Spring Harb Perspect Med. 2017. PMID: 27742734 Free PMC article. Review.
Cited by
-
Antibacterials in Aquatic Environment and Their Toxicity to Fish.Pharmaceuticals (Basel). 2020 Aug 9;13(8):189. doi: 10.3390/ph13080189. Pharmaceuticals (Basel). 2020. PMID: 32784912 Free PMC article. Review.
-
Molecular mechanisms of antibiotic resistance revisited.Nat Rev Microbiol. 2023 May;21(5):280-295. doi: 10.1038/s41579-022-00820-y. Epub 2022 Nov 21. Nat Rev Microbiol. 2023. PMID: 36411397 Review.
-
Antimicrobial resistance in methicillin-resistant Staphylococcus aureus to newer antimicrobial agents.Antimicrob Agents Chemother. 2019 Sep 9;63(12):e01216-19. doi: 10.1128/AAC.01216-19. Epub 2019 Sep 16. Antimicrob Agents Chemother. 2019. PMID: 31527033 Free PMC article. Review.
-
Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors.BMC Vet Res. 2019 May 22;15(1):159. doi: 10.1186/s12917-019-1901-1. BMC Vet Res. 2019. PMID: 31118039 Free PMC article.
-
Maximum levels of cross-contamination for 24 antimicrobial active substances in non-target feed. Part 8: Pleuromutilins: tiamulin and valnemulin.EFSA J. 2021 Oct 26;19(10):e06860. doi: 10.2903/j.efsa.2021.6860. eCollection 2021 Oct. EFSA J. 2021. PMID: 34729088 Free PMC article.
References
-
- Aarestrup FM, Agersø Y, Ahrens P, Jørgensen JC, Madsen M, Jensen LB. 2000. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet Microbiol 74: 353–364. - PubMed
-
- Allignet J, El Solh N. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202: 133–138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
