Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis
- PMID: 27548227
- PMCID: PMC4999840
- DOI: 10.3390/genes7080052
Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis
Abstract
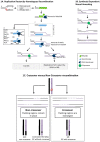
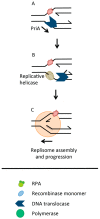
Recombinase enzymes catalyse invasion of single-stranded DNA (ssDNA) into homologous duplex DNA forming "Displacement loops" (D-loops), a process called synapsis. This triggers homologous recombination (HR), which can follow several possible paths to underpin DNA repair and restart of blocked and collapsed DNA replication forks. Therefore, synapsis can be a checkpoint for controlling whether or not, how far, and by which pathway, HR proceeds to overcome an obstacle or break in a replication fork. Synapsis can be antagonized by limiting access of a recombinase to ssDNA and by dissociation of D-loops or heteroduplex formed by synapsis. Antagonists include DNA helicases and translocases that are identifiable in eukaryotes, bacteria and archaea, and which target synaptic and pre-synaptic DNA structures thereby controlling HR at early stages. Here we survey these events with emphasis on enabling DNA replication to be resumed from sites of blockage or collapse. We also note how knowledge of anti-recombination activities could be useful to improve efficiency of CRISPR-based genome editing.
Keywords: Hel308; helicase; homologous recombination; synapsis.
Figures
Similar articles
-
Homologous recombination in Archaea: new Holliday junction helicases.Biochem Soc Trans. 2003 Jun;31(Pt 3):703-5. doi: 10.1042/bst0310703. Biochem Soc Trans. 2003. PMID: 12773187 Review.
-
Recombinases and Related Proteins in the Context of Homologous Recombination Analyzed by Molecular Microscopy.Methods Mol Biol. 2018;1805:251-270. doi: 10.1007/978-1-4939-8556-2_13. Methods Mol Biol. 2018. PMID: 29971722
-
Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks.Mol Cell Biol. 2005 Aug;25(16):7158-69. doi: 10.1128/MCB.25.16.7158-7169.2005. Mol Cell Biol. 2005. PMID: 16055725 Free PMC article.
-
Physical interaction between archaeal DNA repair helicase Hel308 and Replication Protein A (RPA).DNA Repair (Amst). 2011 Mar 7;10(3):306-13. doi: 10.1016/j.dnarep.2010.12.001. Epub 2010 Dec 30. DNA Repair (Amst). 2011. PMID: 21195035
-
The FANCM family of DNA helicases/translocases.DNA Repair (Amst). 2010 Mar 2;9(3):224-36. doi: 10.1016/j.dnarep.2009.12.012. Epub 2010 Feb 8. DNA Repair (Amst). 2010. PMID: 20117061 Review.
Cited by
-
Homologous Recombination as a Fundamental Genome Surveillance Mechanism during DNA Replication.Genes (Basel). 2021 Dec 9;12(12):1960. doi: 10.3390/genes12121960. Genes (Basel). 2021. PMID: 34946909 Free PMC article. Review.
-
BRCA1-A and BRISC: Multifunctional Molecular Machines for Ubiquitin Signaling.Biomolecules. 2020 Oct 31;10(11):1503. doi: 10.3390/biom10111503. Biomolecules. 2020. PMID: 33142801 Free PMC article. Review.
-
Current Implications of microRNAs in Genome Stability and Stress Responses of Ovarian Cancer.Cancers (Basel). 2021 May 29;13(11):2690. doi: 10.3390/cancers13112690. Cancers (Basel). 2021. PMID: 34072593 Free PMC article. Review.
-
Inferring joint sequence-structural determinants of protein functional specificity.Elife. 2018 Jan 16;7:e29880. doi: 10.7554/eLife.29880. Elife. 2018. PMID: 29336305 Free PMC article.
-
Mechanistic and biological considerations of oxidatively damaged DNA for helicase-dependent pathways of nucleic acid metabolism.Free Radic Biol Med. 2017 Jun;107:245-257. doi: 10.1016/j.freeradbiomed.2016.11.022. Epub 2016 Nov 22. Free Radic Biol Med. 2017. PMID: 27884703 Free PMC article. Review.
References
-
- Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
-
- Spies A.M., Kowalczykowski S.C. Homologous recombination by the RecBCD and RecFOR pathways. In: Patrick H.N., editor. The Bacterial Chromosome. ASM Press; Washington, DC, USA: 2005. pp. 398–403.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

