Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults
- PMID: 27546487
- PMCID: PMC4996934
- DOI: 10.1038/ncomms12484
Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults
Abstract
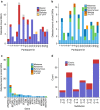
Clonal haematopoiesis is thought to be a rare condition that increases in frequency with age and predisposes individuals to haematological malignancy. Recent studies, utilizing next-generation sequencing (NGS), observed haematopoietic clones in 10% of 70-year olds and rarely in younger individuals. However, these studies could only detect common haematopoietic clones->0.02 variant allele fraction (VAF)-due to the error rate of NGS. To identify and characterize clonal mutations below this threshold, here we develop methods for targeted error-corrected sequencing, which enable the accurate detection of clonal mutations as rare as 0.0003 VAF. We apply these methods to study serially banked peripheral blood samples from healthy 50-60-year-old participants in the Nurses' Health Study. We observe clonal haematopoiesis, frequently harbouring mutations in DNMT3A and TET2, in 95% of individuals studied. These clonal mutations are often stable longitudinally and present in multiple haematopoietic compartments, suggesting a long-lived haematopoietic stem and progenitor cell of origin.
Figures
Similar articles
-
Impact of the variant allele frequency of ASXL1, DNMT3A, JAK2, TET2, TP53, and NPM1 on the outcomes of patients with newly diagnosed acute myeloid leukemia.Cancer. 2020 Feb 15;126(4):765-774. doi: 10.1002/cncr.32566. Epub 2019 Nov 19. Cancer. 2020. PMID: 31742675
-
Clonal haematopoiesis in chronic ischaemic heart failure: prognostic role of clone size for DNMT3A- and TET2-driver gene mutations.Eur Heart J. 2021 Jan 20;42(3):257-265. doi: 10.1093/eurheartj/ehaa845. Eur Heart J. 2021. PMID: 33241418
-
[Clonal haematopoiesis: A concise review].Rev Med Interne. 2019 Oct;40(10):684-692. doi: 10.1016/j.revmed.2019.05.005. Epub 2019 May 22. Rev Med Interne. 2019. PMID: 31126662 Review. French.
-
Clonal hematopoiesis and risk of acute myeloid leukemia.Haematologica. 2019 Dec;104(12):2410-2417. doi: 10.3324/haematol.2018.215269. Epub 2019 Apr 19. Haematologica. 2019. PMID: 31004019 Free PMC article.
-
Current Aspects of Clonal Hematopoiesis: Implications for Clinical Diagnosis.Ann Lab Med. 2019 Nov;39(6):509-514. doi: 10.3343/alm.2019.39.6.509. Ann Lab Med. 2019. PMID: 31240877 Free PMC article. Review.
Cited by
-
Clinical implementation and current advancement of blood liquid biopsy in cancer.J Hum Genet. 2021 Sep;66(9):909-926. doi: 10.1038/s10038-021-00939-5. Epub 2021 Jun 4. J Hum Genet. 2021. PMID: 34088974 Review.
-
High-sensitivity analysis of clonal hematopoiesis reveals increased clonal complexity of potential-driver mutations in severe COVID-19 patients.PLoS One. 2024 Jan 10;19(1):e0282546. doi: 10.1371/journal.pone.0282546. eCollection 2024. PLoS One. 2024. PMID: 38198467 Free PMC article.
-
Clonal Hematopoiesis of Indeterminate Potential as a Novel Risk Factor for Donor-Derived Leukemia.Stem Cell Reports. 2020 Aug 11;15(2):279-291. doi: 10.1016/j.stemcr.2020.07.008. Stem Cell Reports. 2020. PMID: 32783925 Free PMC article. Review.
-
Molecular and clinical aspects relevant for counseling individuals with clonal hematopoiesis of indeterminate potential.Front Oncol. 2023 Dec 15;13:1303785. doi: 10.3389/fonc.2023.1303785. eCollection 2023. Front Oncol. 2023. PMID: 38162500 Free PMC article. Review.
-
Impact of age and sex on myelopoiesis and inflammation during myocardial infarction.J Mol Cell Cardiol. 2024 Feb;187:80-89. doi: 10.1016/j.yjmcc.2023.11.011. Epub 2024 Jan 1. J Mol Cell Cardiol. 2024. PMID: 38163742 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

