Immune cell-specific transcriptional profiling highlights distinct molecular pathways controlled by Tob1 upon experimental autoimmune encephalomyelitis
- PMID: 27546286
- PMCID: PMC4992865
- DOI: 10.1038/srep31603
Immune cell-specific transcriptional profiling highlights distinct molecular pathways controlled by Tob1 upon experimental autoimmune encephalomyelitis
Abstract
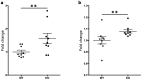
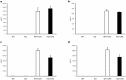
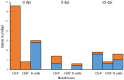
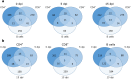
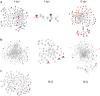
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system characterized by focal lymphocytic infiltration, demyelination and neurodegeneration. Despite the recent advances in understanding MS molecular basis, no reliable biomarkers have been identified yet to monitor disease progression. Our group has previously reported that low levels of TOB1 in CD4(+) T cells are strongly associated with a higher risk of MS conversion in individuals experiencing an initial demyelinating event. Consistently, Tob1 ablation in mice exacerbates the clinical phenotype of the MS model experimental autoimmune encephalomyelitis (EAE). To shed light on Tob1 molecular functions in the immune system, we have conducted the first cell-based transcriptomic analysis in Tob1(-/-) and wildtype mice upon EAE. Next-generation sequencing was employed to characterize the changes in gene expression in T and B cells at pre- and post-symptomatic EAE stages. Remarkably, we found only modest overlap among the different genetic signatures, suggesting that Tob1 may control distinct genetic programs in the different cytotypes. This hypothesis was corroborated by gene ontology and global interactome analyses, which highlighted specific cellular pathways in each cellular subset before and after EAE induction. In summary, our work pinpoints a multifaceted activity of Tob1 in both homeostasis and disease progression.
Figures

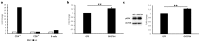
Similar articles
-
Tob1 plays a critical role in the activation of encephalitogenic T cells in CNS autoimmunity.J Exp Med. 2013 Jul 1;210(7):1301-9. doi: 10.1084/jem.20121611. J Exp Med. 2013. PMID: 23797093 Free PMC article.
-
B cells are capable of independently eliciting rapid reactivation of encephalitogenic CD4 T cells in a murine model of multiple sclerosis.PLoS One. 2018 Jun 26;13(6):e0199694. doi: 10.1371/journal.pone.0199694. eCollection 2018. PLoS One. 2018. PMID: 29944721 Free PMC article.
-
Emerging role of IL-16 in cytokine-mediated regulation of multiple sclerosis.Cytokine. 2015 Oct;75(2):234-48. doi: 10.1016/j.cyto.2015.01.005. Epub 2015 Feb 18. Cytokine. 2015. PMID: 25703787
-
Emerging concepts in autoimmune encephalomyelitis beyond the CD4/T(H)1 paradigm.Ann Anat. 2010 Aug 20;192(4):179-93. doi: 10.1016/j.aanat.2010.06.006. Epub 2010 Jul 15. Ann Anat. 2010. PMID: 20692821 Review.
-
What transgenic and knockout mouse models teach us about experimental autoimmune encephalomyelitis.Rev Immunogenet. 2000;2(1):115-32. Rev Immunogenet. 2000. PMID: 11324684 Review.
Cited by
-
Association between the SNPs of the TOB1 gene and gastric cancer risk in the Chinese Han population of northeast China.J Cancer. 2018 Apr 6;9(8):1371-1378. doi: 10.7150/jca.23805. eCollection 2018. J Cancer. 2018. PMID: 29721046 Free PMC article.
-
Hyperpolarized 13C MR metabolic imaging can detect neuroinflammation in vivo in a multiple sclerosis murine model.Proc Natl Acad Sci U S A. 2017 Aug 15;114(33):E6982-E6991. doi: 10.1073/pnas.1613345114. Epub 2017 Jul 31. Proc Natl Acad Sci U S A. 2017. PMID: 28760957 Free PMC article.
-
A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis.Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):22932-22943. doi: 10.1073/pnas.2008523117. Epub 2020 Aug 28. Proc Natl Acad Sci U S A. 2020. PMID: 32859762 Free PMC article.
-
TOB1 modulates neutrophil phenotypes to influence gastric cancer progression and immunotherapy efficacy.Front Immunol. 2024 Mar 28;15:1369087. doi: 10.3389/fimmu.2024.1369087. eCollection 2024. Front Immunol. 2024. PMID: 38617839 Free PMC article.
-
Sex-specific Tau methylation patterns and synaptic transcriptional alterations are associated with neural vulnerability during chronic neuroinflammation.J Autoimmun. 2019 Jul;101:56-69. doi: 10.1016/j.jaut.2019.04.003. Epub 2019 Apr 19. J Autoimmun. 2019. PMID: 31010726 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

