PML at Mitochondria-Associated Membranes Is Critical for the Repression of Autophagy and Cancer Development
- PMID: 27545895
- PMCID: PMC5011426
- DOI: 10.1016/j.celrep.2016.07.082
PML at Mitochondria-Associated Membranes Is Critical for the Repression of Autophagy and Cancer Development
Abstract
The precise molecular mechanisms that coordinate apoptosis and autophagy in cancer remain to be determined. Here, we provide evidence that the tumor suppressor promyelocytic leukemia protein (PML) controls autophagosome formation at mitochondria-associated membranes (MAMs) and, thus, autophagy induction. Our in vitro and in vivo results demonstrate how PML functions as a repressor of autophagy. PML loss promotes tumor development, providing a growth advantage to tumor cells that use autophagy as a cell survival strategy during stress conditions. These findings demonstrate that autophagy inhibition could be paired with a chemotherapeutic agent to develop anticancer strategies for tumors that present PML downregulation.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
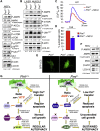
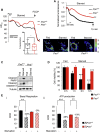
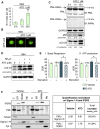
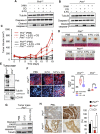
Comment in
-
Novel Insights into PML-Dependent Oncosuppression.Trends Cell Biol. 2016 Dec;26(12):889-890. doi: 10.1016/j.tcb.2016.09.001. Epub 2016 Sep 20. Trends Cell Biol. 2016. PMID: 27663133
Similar articles
-
TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of p53-Mediated Senescence.Cancer Cell. 2017 May 8;31(5):697-710.e7. doi: 10.1016/j.ccell.2017.04.006. Cancer Cell. 2017. PMID: 28486108
-
The arsenic-based cure of acute promyelocytic leukemia promotes cytoplasmic sequestration of PML and PML/RARA through inhibition of PML body recycling.Blood. 2012 Jul 26;120(4):847-57. doi: 10.1182/blood-2011-10-388496. Epub 2012 Jun 12. Blood. 2012. PMID: 22692509
-
Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein.Blood. 2010 Sep 30;116(13):2324-31. doi: 10.1182/blood-2010-01-261040. Epub 2010 Jun 23. Blood. 2010. PMID: 20574048
-
The role of PML in hematopoietic and leukemic stem cell maintenance.Int J Hematol. 2014 Jul;100(1):18-26. doi: 10.1007/s12185-014-1518-x. Epub 2014 Feb 1. Int J Hematol. 2014. PMID: 24488785 Free PMC article. Review.
-
The functional roles of PML nuclear bodies in genome maintenance.Mutat Res. 2018 May;809:99-107. doi: 10.1016/j.mrfmmm.2017.05.002. Epub 2017 May 5. Mutat Res. 2018. PMID: 28521962 Review.
Cited by
-
Defective endoplasmic reticulum-mitochondria contacts and bioenergetics in SEPN1-related myopathy.Cell Death Differ. 2021 Jan;28(1):123-138. doi: 10.1038/s41418-020-0587-z. Epub 2020 Jul 13. Cell Death Differ. 2021. PMID: 32661288 Free PMC article.
-
Mitochondrial P2X7 Receptor Localization Modulates Energy Metabolism Enhancing Physical Performance.Function (Oxf). 2021 Jan 28;2(2):zqab005. doi: 10.1093/function/zqab005. eCollection 2021. Function (Oxf). 2021. PMID: 35330818 Free PMC article.
-
Autophagy in the Neuronal Ceroid Lipofuscinoses (Batten Disease).Front Cell Dev Biol. 2022 Feb 16;10:812728. doi: 10.3389/fcell.2022.812728. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35252181 Free PMC article. Review.
-
Understanding the Role of Autophagy in Cancer Formation and Progression Is a Real Opportunity to Treat and Cure Human Cancers.Cancers (Basel). 2021 Nov 10;13(22):5622. doi: 10.3390/cancers13225622. Cancers (Basel). 2021. PMID: 34830777 Free PMC article. Review.
-
Dcf1 induces glioblastoma cells apoptosis by blocking autophagy.Cancer Med. 2022 Jan;11(1):207-223. doi: 10.1002/cam4.4440. Epub 2021 Nov 19. Cancer Med. 2022. PMID: 34799992 Free PMC article.
References
-
- Ablain J., Rice K., Soilihi H., de Reynies A., Minucci S., de Thé H. Activation of a promyelocytic leukemia-tumor protein 53 axis underlies acute promyelocytic leukemia cure. Nat. Med. 2014;20:167–174. - PubMed
-
- Bonora M., Giorgi C., Bononi A., Marchi S., Patergnani S., Rimessi A., Rizzuto R., Pinton P. Subcellular calcium measurements in mammalian cells using jellyfish photoprotein aequorin-based probes. Nat. Protoc. 2013;8:2105–2118. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

