miR-20a regulates proliferation, differentiation and apoptosis in P19 cell model of cardiac differentiation by targeting Smoothened
- PMID: 27543062
- PMCID: PMC5051645
- DOI: 10.1242/bio.019182
miR-20a regulates proliferation, differentiation and apoptosis in P19 cell model of cardiac differentiation by targeting Smoothened
Abstract
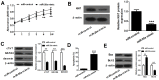
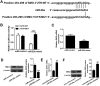
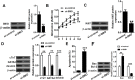
MicroRNA (miR)-20a, a member of the miR-17-92 cluster related to cardiac development, was obviously downregulated in myocardially differentiated P19 cells compared with normal P19 cells. Smoothened (SMO) is a member of the Hh pathway. Hh signaling induces cardiac differentiation in P19 cells, and SMO mediates the Hh pathway during embryonic development. Using bioinformatic prediction software Targetscan (http://www.targetscan.org/), PicTar (http://pictar.bio.nyu.edu), and miRBase (http://microrna.sanger.ac.uk/), miR-20a and the 3'-untranslated region (3'-UTR) of SMO mRNA were predicted to have complementary binding regions. Accordingly, we inferred that miR-20a might act as a regulator of SMO, and regulate proliferation, differentiation and apoptosis in P19 cells. We determined the expression of miR-20a, SMO and marker proteins of cardiomyocytes (cTnT, GATA4 and desmin) by quantitative real-time PCR (qRT-PCR) and western blot assays, and found that P19 cells had differentiated into cardiomyocytes successfully at differentiation day 10, and downregulation of miR-20a and upregulation of SMO existed in myocardially differentiated P19 cells. Cell proliferation, differentiation and apoptosis detection showed that miR-20a upregulation inhibited proliferation and differentiation and enhanced apoptosis in P19 cells. Moreover, we verified that miR-20a directly targeted SMO and knockdown of SMO and miR-20a overexpression had similar effects on P19 cell proliferation, differentiation and apoptosis, which verified the speculation that miR-20a inhibits proliferation and differentiation and enhances apoptosis in P19 cells by directly targeting SMO. Our results suggest that miR-20a may be a potential target against congenital heart diseases.
Keywords: Apoptosis; Differentiation; P19 cell; Proliferation; Smoothened; miR-20a.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
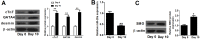
Similar articles
-
Effect of miR-20b on Apoptosis, Differentiation, the BMP Signaling Pathway and Mitochondrial Function in the P19 Cell Model of Cardiac Differentiation In Vitro.PLoS One. 2015 Apr 21;10(4):e0123519. doi: 10.1371/journal.pone.0123519. eCollection 2015. PLoS One. 2015. PMID: 25898012 Free PMC article.
-
Effects of miR-19b overexpression on proliferation, differentiation, apoptosis and Wnt/β-catenin signaling pathway in P19 cell model of cardiac differentiation in vitro.Cell Biochem Biophys. 2013 Jul;66(3):709-22. doi: 10.1007/s12013-013-9516-9. Cell Biochem Biophys. 2013. PMID: 23443808
-
[miR-25-3p down-regulates the expression of ADAM10 to inhibit the differentiation of P19 cells into cardiomyocytes by blocking the Notch signaling pathway].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2019 May;35(5):405-411. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2019. PMID: 31223109 Chinese.
-
MicroRNA-29c overexpression inhibits proliferation and promotes apoptosis and differentiation in P19 embryonal carcinoma cells.Gene. 2016 Jan 15;576(1 Pt 2):304-11. doi: 10.1016/j.gene.2015.10.038. Epub 2015 Oct 19. Gene. 2016. PMID: 26484393
-
Restoration of miR-98 relieves the inhibitory effect of nicotine on the differentiation of P19 cells into cardiomyocytes.Biotechnol Lett. 2016 Apr;38(4):579-87. doi: 10.1007/s10529-015-2030-y. Epub 2015 Dec 31. Biotechnol Lett. 2016. PMID: 26721233
Cited by
-
Epigenetic Modification Factors and microRNAs Network Associated with Differentiation of Embryonic Stem Cells and Induced Pluripotent Stem Cells toward Cardiomyocytes: A Review.Life (Basel). 2023 Feb 17;13(2):569. doi: 10.3390/life13020569. Life (Basel). 2023. PMID: 36836926 Free PMC article. Review.
-
The miR-17-92 cluster in cardiac health and disease.Birth Defects Res. 2024 Jan;116(1):e2273. doi: 10.1002/bdr2.2273. Epub 2023 Nov 20. Birth Defects Res. 2024. PMID: 37984445 Free PMC article. Review.
-
TMAO Upregulates Members of the miR-17/92 Cluster and Impacts Targets Associated with Atherosclerosis.Int J Mol Sci. 2022 Oct 11;23(20):12107. doi: 10.3390/ijms232012107. Int J Mol Sci. 2022. PMID: 36292963 Free PMC article.
-
Relations between plasma microRNAs, echocardiographic markers of atrial remodeling, and atrial fibrillation: Data from the Framingham Offspring study.PLoS One. 2020 Aug 19;15(8):e0236960. doi: 10.1371/journal.pone.0236960. eCollection 2020. PLoS One. 2020. PMID: 32813736 Free PMC article.
-
Insights into role of microRNAs in cardiac development, cardiac diseases, and developing novel therapies.Iran J Basic Med Sci. 2020 Aug;23(8):961-969. doi: 10.22038/ijbms.2020.40974.10015. Iran J Basic Med Sci. 2020. PMID: 32952941 Free PMC article. Review.
References
-
- Capozzi G., Caputo S., Pizzuti R., Martina L., Santoro M., Santoro G., Sarubbi B., Iacono C., D'Alto M. and Bigazzi M. C. (2008). Congenital heart disease in live-born children: incidence, distribution, and yearly changes in the Campania Region. J. Cardiovasc. Med. 9, 368-374. 10.2459/JCM.0b013e3282eee866 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

