Solid-state NMR characterization of amphomycin effects on peptidoglycan and wall teichoic acid biosyntheses in Staphylococcus aureus
- PMID: 27538449
- PMCID: PMC4990924
- DOI: 10.1038/srep31757
Solid-state NMR characterization of amphomycin effects on peptidoglycan and wall teichoic acid biosyntheses in Staphylococcus aureus
Abstract
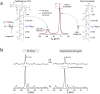
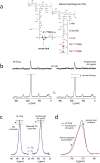
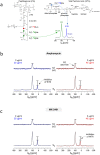
Amphomycin and MX-2401 are cyclic lipopeptides exhibiting bactericidal activities against Gram-positive pathogens. Amphomycin and MX-2401 share structural similarities with daptomycin, but unlike daptomycin they do not target bacterial membrane. In this study, we investigate in vivo modes of action for amphomycin and MX-2401 in intact whole cells of Staphylococcus aureus by measuring the changes of peptidoglycan and wall teichoic acid compositions using solid-state NMR. S. aureus were grown in a defined media containing isotope labels [1-(13)C]glycine and L-[ε-(15)N]lysin, L-[1-(13)C]lysine and D-[(15)N]alanine, or D-[1-(13)C]alanine and [(15)N]glycine, to selectively (13)C-(15)N pair label peptidoglycan bridge-link, stem-link, and cross-link, respectively. (13)C{(15)N} and (15)N{(13)C} rotational-echo double resonance NMR measurements determined that cyclic lipopeptide-treated S. aureus exhibited thinning of the cell wall, accumulation of Park's nucleotide, inhibition of glycine utilization for purine biosynthesis, reduction of ester-linked D-Ala in teichoic acids, and reduction of peptidoglycan cross-linking. Whole cell NMR analysis also revealed that S. aureus, in presence of amphomycin and MX-2401, maintained the incorporation of D-Ala during peptidoglycan biosynthesis while the incorporation of D-Ala into teichoic acids was inhibited. These effects are consistent with amphomycin's dual inhibition of both peptidoglycan and wall teichoic acid biosyntheses in S. aureus.
Figures

Similar articles
-
Hidden Mode of Action of Glycopeptide Antibiotics: Inhibition of Wall Teichoic Acid Biosynthesis.J Phys Chem B. 2017 Apr 27;121(16):3925-3932. doi: 10.1021/acs.jpcb.7b00324. Epub 2017 Apr 14. J Phys Chem B. 2017. PMID: 28368603 Free PMC article.
-
Conformational and quantitative characterization of oritavancin-peptidoglycan complexes in whole cells of Staphylococcus aureus by in vivo 13C and 15N labeling.J Mol Biol. 2006 Apr 7;357(4):1253-62. doi: 10.1016/j.jmb.2006.01.040. Epub 2006 Jan 30. J Mol Biol. 2006. PMID: 16483598
-
Peptidoglycan and Teichoic Acid Levels and Alterations in Staphylococcus aureus by Cell-Wall and Whole-Cell Nuclear Magnetic Resonance.Biochemistry. 2018 Jul 3;57(26):3966-3975. doi: 10.1021/acs.biochem.8b00495. Epub 2018 Jun 11. Biochemistry. 2018. PMID: 29806458 Free PMC article.
-
Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics.J Antibiot (Tokyo). 2014 Jan;67(1):43-51. doi: 10.1038/ja.2013.100. Epub 2013 Oct 30. J Antibiot (Tokyo). 2014. PMID: 24169797 Review.
-
Occurrence and function of membrane teichoic acids.Biochim Biophys Acta. 1977 May 31;472(1):1-12. doi: 10.1016/0304-4157(77)90012-0. Biochim Biophys Acta. 1977. PMID: 406922 Review.
Cited by
-
Chemical Biology Tools for Examining the Bacterial Cell Wall.Cell Chem Biol. 2020 Aug 20;27(8):1052-1062. doi: 10.1016/j.chembiol.2020.07.024. Cell Chem Biol. 2020. PMID: 32822617 Free PMC article. Review.
-
Crystallomycin revisited after 60 years: aspartocins B and C.Medchemcomm. 2018 Feb 27;9(4):667-675. doi: 10.1039/c8md00002f. eCollection 2018 Apr 1. Medchemcomm. 2018. PMID: 30108957 Free PMC article.
-
The calcium-dependent lipopeptide antibiotics: structure, mechanism, & medicinal chemistry.Medchemcomm. 2019 Mar 21;10(5):634-646. doi: 10.1039/c9md00126c. eCollection 2019 May 1. Medchemcomm. 2019. PMID: 31191855 Free PMC article. Review.
-
An Overview of Microorganisms Immobilized in a Gel Structure for the Production of Precursors, Antibiotics, and Valuable Products.Gels. 2024 Oct 10;10(10):646. doi: 10.3390/gels10100646. Gels. 2024. PMID: 39451299 Free PMC article. Review.
-
Bioinformatic prospecting and synthesis of a bifunctional lipopeptide antibiotic that evades resistance.Science. 2022 May 27;376(6596):991-996. doi: 10.1126/science.abn4213. Epub 2022 May 26. Science. 2022. PMID: 35617397 Free PMC article.
References
-
- Tanaka H. et al.. Studies on bacterial cell wall inhibitors. II. Inhibition of peptidoglycan synthesis in vivo and in vitro by amphomycin. Biochim Biophys Acta 497, 633–640 (1977). - PubMed
-
- Hu Y., Helm J. S., Chen L., Ye X. Y. & Walker S. Ramoplanin inhibits bacterial transglycosylases by binding as a dimer to lipid II. J Am Chem Soc 125, 8736–8737 (2003). - PubMed
-
- Kato A. et al.. A new anti-MRSA antibiotic complex, WAP-8294A. I. Taxonomy, isolation and biological activities. J Antibiot (Tokyo) 51, 929–935 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

