Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold
- PMID: 27528683
- PMCID: PMC5024648
- DOI: 10.1073/pnas.1605031113
Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold
Abstract
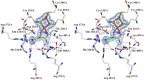
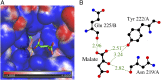
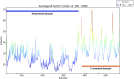
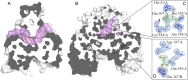
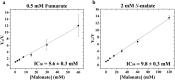
Fumarate hydratases (FHs) are essential metabolic enzymes grouped into two classes. Here, we present the crystal structure of a class I FH, the cytosolic FH from Leishmania major, which reveals a previously undiscovered protein fold that coordinates a catalytically essential [4Fe-4S] cluster. Our 2.05 Å resolution data further reveal a dimeric architecture for this FH that resembles a heart, with each lobe comprised of two domains that are arranged around the active site. Besides the active site, where the substrate S-malate is bound bidentate to the unique iron of the [4Fe-4S] cluster, other binding pockets are found near the dimeric enzyme interface, some of which are occupied by malonate, shown here to be a weak inhibitor of this enzyme. Taken together, these data provide a framework both for investigations of the class I FH catalytic mechanism and for drug design aimed at fighting neglected tropical diseases.
Keywords: Fe-S cluster; X-ray crystallography; fumarate hydratase; leishmaniases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
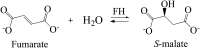


Similar articles
-
Structural and Biochemical Investigations of the [4Fe-4S] Cluster-Containing Fumarate Hydratase from Leishmania major.Biochemistry. 2019 Dec 10;58(49):5011-5021. doi: 10.1021/acs.biochem.9b00923. Epub 2019 Nov 27. Biochemistry. 2019. PMID: 31743022 Free PMC article.
-
Crystallographic snapshots of sulfur insertion by lipoyl synthase.Proc Natl Acad Sci U S A. 2016 Aug 23;113(34):9446-50. doi: 10.1073/pnas.1602486113. Epub 2016 Aug 9. Proc Natl Acad Sci U S A. 2016. PMID: 27506792 Free PMC article.
-
Crystal Structures of Fumarate Hydratases from Leishmania major in a Complex with Inhibitor 2-Thiomalate.ACS Chem Biol. 2019 Feb 15;14(2):266-275. doi: 10.1021/acschembio.8b00972. Epub 2019 Jan 24. ACS Chem Biol. 2019. PMID: 30645090 Free PMC article.
-
Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.Acta Crystallogr F Struct Biol Commun. 2018 Oct 1;74(Pt 10):617-624. doi: 10.1107/S2053230X18011895. Epub 2018 Sep 19. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 30279312 Free PMC article.
-
Trapping a cross-linked lysine-tryptophan radical in the catalytic cycle of the radical SAM enzyme SuiB.Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2101571118. doi: 10.1073/pnas.2101571118. Proc Natl Acad Sci U S A. 2021. PMID: 34001621 Free PMC article.
Cited by
-
Structural and Biochemical Investigations of the [4Fe-4S] Cluster-Containing Fumarate Hydratase from Leishmania major.Biochemistry. 2019 Dec 10;58(49):5011-5021. doi: 10.1021/acs.biochem.9b00923. Epub 2019 Nov 27. Biochemistry. 2019. PMID: 31743022 Free PMC article.
-
Biochemical characterization and essentiality of Plasmodium fumarate hydratase.J Biol Chem. 2018 Apr 20;293(16):5878-5894. doi: 10.1074/jbc.M117.816298. Epub 2018 Feb 15. J Biol Chem. 2018. PMID: 29449371 Free PMC article.
-
Introducing CRAFT: The Center for Research and Advancement in Fragments and molecular Targets.ACS Med Chem Lett. 2024 Jul 23;15(8):1174-1177. doi: 10.1021/acsmedchemlett.4c00296. eCollection 2024 Aug 8. ACS Med Chem Lett. 2024. PMID: 39140068 Free PMC article.
-
Chemoproteomics Reveals Disruption of Metal Homeostasis and Metalloproteins by the Antibiotic Holomycin.ACS Chem Biol. 2023 Sep 15;18(9):1909-1914. doi: 10.1021/acschembio.3c00360. Epub 2023 Aug 10. ACS Chem Biol. 2023. PMID: 37561838 Free PMC article.
-
Aurothiomalate-Based Drugs as Potentially Novel Agents Against Leishmania major: A Mini Review.Acta Parasitol. 2022 Jun;67(2):640-647. doi: 10.1007/s11686-022-00536-2. Epub 2022 Apr 5. Acta Parasitol. 2022. PMID: 35380401 Review.
References
-
- Coustou V, et al. A mitochondrial NADH-dependent fumarate reductase involved in the production of succinate excreted by procyclic Trypanosoma brucei. J Biol Chem. 2005;280(17):16559–16570. - PubMed
-
- Cordeiro AT, Feliciano PR, Pinheiro MP, Nonato MC. Crystal structure of dihydroorotate dehydrogenase from Leishmania major. Biochimie. 2012;94(8):1739–1748. - PubMed
-
- Coustou V, et al. Fumarate is an essential intermediary metabolite produced by the procyclic Trypanosoma brucei. J Biol Chem. 2006;281(37):26832–26846. - PubMed
-
- Feliciano PR, et al. Fumarate hydratase isoforms of Leishmania major: Subcellular localization, structural and kinetic properties. Int J Biol Macromol. 2012;51(1-2):25–31. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

