Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm
- PMID: 27521372
- PMCID: PMC5062992
- DOI: 10.1093/nar/gkw723
Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm
Abstract
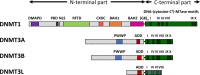
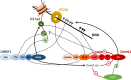
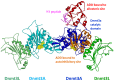
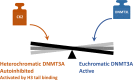
In mammals, DNA methylation is introduced by the DNMT1, DNMT3A and DNMT3B methyltransferases, which are all large multi-domain proteins containing a catalytic C-terminal domain and an N-terminal part with regulatory functions. Recently, two novel regulatory principles of DNMTs were uncovered. It was shown that their catalytic activity is under allosteric control of N-terminal domains with autoinhibitory function, the RFT and CXXC domains in DNMT1 and the ADD domain in DNMT3. Moreover, targeting and activity of DNMTs were found to be regulated in a concerted manner by interactors and posttranslational modifications (PTMs). In this review, we describe the structures and domain composition of the DNMT1 and DNMT3 enzymes, their DNA binding, catalytic mechanism, multimerization and the processes controlling their stability in cells with a focus on their regulation and chromatin targeting by PTMs, interactors and chromatin modifications. We propose that the allosteric regulation of DNMTs by autoinhibitory domains acts as a general switch for the modulation of the function of DNMTs, providing numerous possibilities for interacting proteins, nucleic acids or PTMs to regulate DNMT activity and targeting. The combined regulation of DNMT targeting and catalytic activity contributes to the precise spatiotemporal control of DNMT function and genome methylation in cells.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
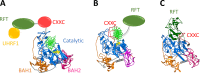

Similar articles
-
Enzymology of Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2016;945:87-122. doi: 10.1007/978-3-319-43624-1_5. Adv Exp Med Biol. 2016. PMID: 27826836 Review.
-
Enzymology of Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2022;1389:69-110. doi: 10.1007/978-3-031-11454-0_4. Adv Exp Med Biol. 2022. PMID: 36350507
-
Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells.Prog Mol Biol Transl Sci. 2011;101:311-33. doi: 10.1016/B978-0-12-387685-0.00009-3. Prog Mol Biol Transl Sci. 2011. PMID: 21507356 Review.
-
The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA.J Mol Biol. 2001 Jun 22;309(5):1189-99. doi: 10.1006/jmbi.2001.4709. J Mol Biol. 2001. PMID: 11399088
-
Interactions within the mammalian DNA methyltransferase family.BMC Mol Biol. 2003 May 30;4:7. doi: 10.1186/1471-2199-4-7. Epub 2003 May 30. BMC Mol Biol. 2003. PMID: 12777184 Free PMC article.
Cited by
-
Inference of DNA methylation patterns in molluscs.Philos Trans R Soc Lond B Biol Sci. 2021 May 24;376(1825):20200166. doi: 10.1098/rstb.2020.0166. Epub 2021 Apr 5. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33813896 Free PMC article.
-
Moderate DNA hypomethylation suppresses intestinal tumorigenesis by promoting caspase-3 expression and apoptosis.Oncogenesis. 2021 May 4;10(5):38. doi: 10.1038/s41389-021-00328-9. Oncogenesis. 2021. PMID: 33947834 Free PMC article.
-
The DNMT3A R882H mutation does not cause dominant negative effects in purified mixed DNMT3A/R882H complexes.Sci Rep. 2018 Sep 5;8(1):13242. doi: 10.1038/s41598-018-31635-8. Sci Rep. 2018. PMID: 30185810 Free PMC article.
-
Methylation of recombinant mononucleosomes by DNMT3A demonstrates efficient linker DNA methylation and a role of H3K36me3.Commun Biol. 2022 Mar 2;5(1):192. doi: 10.1038/s42003-022-03119-z. Commun Biol. 2022. PMID: 35236925 Free PMC article.
-
Both intra and inter-domain interactions define the intrinsic dynamics and allosteric mechanism in DNMT1s.Comput Struct Biotechnol J. 2020 Mar 23;18:749-764. doi: 10.1016/j.csbj.2020.03.016. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32280430 Free PMC article.
References
-
- Jeltsch A., Jurkowska R.Z. New concepts in DNA methylation. Trends Biochem. Sci. 2014;39:310–318. - PubMed
-
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. - PubMed
-
- Jurkowska R.Z., Jurkowski T.P., Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. - PubMed
-
- Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. - PubMed
-
- Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

