Conditional Disruption of Raptor Reveals an Essential Role for mTORC1 in B Cell Development, Survival, and Metabolism
- PMID: 27521345
- PMCID: PMC5009877
- DOI: 10.4049/jimmunol.1600492
Conditional Disruption of Raptor Reveals an Essential Role for mTORC1 in B Cell Development, Survival, and Metabolism
Abstract
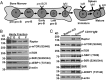
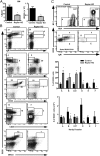
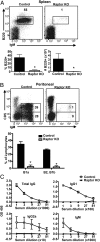
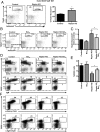
Mechanistic target of rapamycin (mTOR) is a serine-threonine kinase that coordinates nutrient and growth factor availability with cellular growth, division, and differentiation. Studies examining the roles of mTOR signaling in immune function revealed critical roles for mTOR in regulating T cell differentiation and function. However, few studies have investigated the roles of mTOR in early B cell development. In this study, we found that mTOR is highly activated during the pro- and pre-B stages of mouse B cell development. Conditional disruption of the mTOR coactivating protein Raptor in developing mouse B cells resulted in a developmental block at the pre-B cell stage, with a corresponding lack of peripheral B cells and loss of Ag-specific Ab production. Pre-B cell survival and proliferation were significantly reduced in Raptor-deficient mice. Forced expression of a transgenic BCR or a BclxL transgene on Raptor-deficient B cells failed to rescue B cell development, suggesting that pre-BCR signaling and B cell survival are impaired in a BclxL-independent manner. Raptor-deficient pre-B cells exhibited significant decreases in oxidative phosphorylation and glycolysis, indicating that loss of mTOR signaling in B cells significantly impairs cellular metabolic capacity. Treatment of mice with rapamycin, an allosteric inhibitor of mTOR, recapitulated the early B cell developmental block. Collectively, our data reveal a previously uncharacterized role for mTOR signaling in early B cell development, survival, and metabolism.
Copyright © 2016 by The American Association of Immunologists, Inc.
Figures


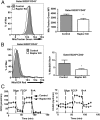
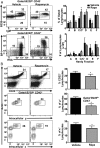
Similar articles
-
Mammalian target of rapamycin complex 1 signalling is essential for germinal centre reaction.Immunology. 2017 Oct;152(2):276-286. doi: 10.1111/imm.12767. Epub 2017 Jul 10. Immunology. 2017. PMID: 28557002 Free PMC article.
-
Synergistic Effects between mTOR Complex 1/2 and Glycolysis Inhibitors in Non-Small-Cell Lung Carcinoma Cells.PLoS One. 2015 Jul 15;10(7):e0132880. doi: 10.1371/journal.pone.0132880. eCollection 2015. PLoS One. 2015. PMID: 26176608 Free PMC article.
-
mTOR has a developmental stage-specific role in mitochondrial fitness independent of conventional mTORC1 and mTORC2 and the kinase activity.PLoS One. 2017 Aug 16;12(8):e0183266. doi: 10.1371/journal.pone.0183266. eCollection 2017. PLoS One. 2017. PMID: 28813526 Free PMC article.
-
The Role of Mammalian Target of Rapamycin (mTOR) in Insulin Signaling.Nutrients. 2017 Oct 27;9(11):1176. doi: 10.3390/nu9111176. Nutrients. 2017. PMID: 29077002 Free PMC article. Review.
-
Raptor and mTOR: subunits of a nutrient-sensitive complex.Curr Top Microbiol Immunol. 2004;279:259-70. doi: 10.1007/978-3-642-18930-2_15. Curr Top Microbiol Immunol. 2004. PMID: 14560962 Review.
Cited by
-
Folliculin Interacting Protein 1 Maintains Metabolic Homeostasis during B Cell Development by Modulating AMPK, mTORC1, and TFE3.J Immunol. 2019 Dec 1;203(11):2899-2908. doi: 10.4049/jimmunol.1900395. Epub 2019 Nov 1. J Immunol. 2019. PMID: 31676673 Free PMC article.
-
The role of mTOR-mediated signals during haemopoiesis and lineage commitment.Biochem Soc Trans. 2018 Oct 19;46(5):1313-1324. doi: 10.1042/BST20180141. Epub 2018 Aug 28. Biochem Soc Trans. 2018. PMID: 30154096 Free PMC article. Review.
-
Reduced eIF4E function impairs B-cell leukemia without altering normal B-lymphocyte function.iScience. 2021 Jun 17;24(7):102748. doi: 10.1016/j.isci.2021.102748. eCollection 2021 Jul 23. iScience. 2021. PMID: 34278258 Free PMC article.
-
The multifaceted role of autophagy in skin autoimmune disorders: a guardian or culprit?Front Immunol. 2024 Apr 16;15:1343987. doi: 10.3389/fimmu.2024.1343987. eCollection 2024. Front Immunol. 2024. PMID: 38690268 Free PMC article. Review.
-
mTORC1 activity is essential for erythropoiesis and B cell lineage commitment.Sci Rep. 2019 Nov 15;9(1):16917. doi: 10.1038/s41598-019-53141-1. Sci Rep. 2019. PMID: 31729420 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

