Epigenetic blockade of neoplastic transformation by bromodomain and extra-terminal (BET) domain protein inhibitor JQ-1
- PMID: 27520485
- PMCID: PMC5031540
- DOI: 10.1016/j.bcp.2016.08.009
Epigenetic blockade of neoplastic transformation by bromodomain and extra-terminal (BET) domain protein inhibitor JQ-1
Abstract
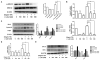
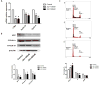
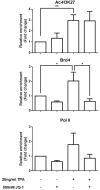
The neoplastic transformation of cells and inflammation are processes that contribute to tumor initiation. Recently, emerging evidence has suggested that epigenetic alterations are also implicated in the early stages of carcinogenesis. Therefore, potent small molecules targeting epigenetic regulators have been developed as novel cancer therapeutic and preventive strategies. Bromodomain and extraterminal domain (BET) proteins are epigenetic readers that play key roles at the interface between chromatin modification and transcriptional regulation. In this study, we investigated the effect of the BET inhibitor JQ-1 on malignant transformation induced by 12-O-tetradecanoylphorbol-13-acetate (TPA) in mouse skin epidermal JB6 P+ cells. Treatment with JQ-1 effectively impaired TPA-induced colony formation in vitro. At the molecular level, the expression of several key TPA-induced pro-survival and pro-proliferative genes (Bcl2, Cyclin D1, and c-Myc) decreased rapidly after BET inhibition. In addition, JQ-1 treatment attenuated the activation of inflammatory NF-κB signaling triggered by TPA. Luciferase reporter assays using plasmids carrying different elements from the COX2 or IL6 promoters demonstrated that JQ-1 does not directly inhibit interactions between NF-κB and its binding sequence; rather, it affects CRE-element-associated transcriptional enhancement. Through siRNA gene silencing, we found that JQ-1 inhibits the p300-dependent transcriptional activation of COX2, which correlates with the results of the luciferase assay. Chromatin immunoprecipitation assays showed that TPA elevated H3K27Ac enrichment in the COX2 promoter region, which is mediated by p300, and Brd4. JQ-1 treatment did not change H3K27Ac levels but decreased the recruitment of Brd4 and RNA Polymerase II. Collectively, our study reveals that the BET inhibitor JQ-1 exerts potent anti-cancer and anti-inflammatory effects by interfering with the core transcriptional program of neoplastic transformation.
Keywords: Brd4; Bromodomain; JQ-1; NF-κB; Neoplastic transformation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
of potential conflicts of interest: No potential conflicts of interest were disclosed.
Figures

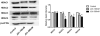

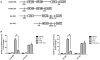

Similar articles
-
Involvement of the antioxidative property of morusin in blocking phorbol ester-induced malignant transformation of JB6 P+ mouse epidermal cells.Chem Biol Interact. 2017 Feb 25;264:34-42. doi: 10.1016/j.cbi.2017.01.009. Epub 2017 Jan 17. Chem Biol Interact. 2017. PMID: 28108223
-
Disruption of BRD4 at H3K27Ac-enriched enhancer region correlates with decreased c-Myc expression in Merkel cell carcinoma.Epigenetics. 2015;10(6):460-6. doi: 10.1080/15592294.2015.1034416. Epub 2015 May 5. Epigenetics. 2015. PMID: 25941994 Free PMC article.
-
Bromodomain and extraterminal (BET) protein inhibition suppresses human T cell leukemia virus 1 (HTLV-1) Tax protein-mediated tumorigenesis by inhibiting nuclear factor κB (NF-κB) signaling.J Biol Chem. 2013 Dec 13;288(50):36094-105. doi: 10.1074/jbc.M113.485029. Epub 2013 Nov 4. J Biol Chem. 2013. PMID: 24189064 Free PMC article.
-
BET Bromodomain as a Target of Epigenetic Therapy.Chem Pharm Bull (Tokyo). 2016;64(6):540-7. doi: 10.1248/cpb.c16-00225. Chem Pharm Bull (Tokyo). 2016. PMID: 27250788 Review.
-
BET bromodomain inhibitors--a novel epigenetic approach in castration-resistant prostate cancer.Cancer Biol Ther. 2014;15(12):1583-5. doi: 10.4161/15384047.2014.962297. Cancer Biol Ther. 2014. PMID: 25535892 Free PMC article. Review.
Cited by
-
The BET family in immunity and disease.Signal Transduct Target Ther. 2021 Jan 19;6(1):23. doi: 10.1038/s41392-020-00384-4. Signal Transduct Target Ther. 2021. PMID: 33462181 Free PMC article. Review.
-
Emerging roles of and therapeutic strategies targeting BRD4 in cancer.Cell Immunol. 2019 Mar;337:48-53. doi: 10.1016/j.cellimm.2019.02.001. Epub 2019 Feb 4. Cell Immunol. 2019. PMID: 30832981 Free PMC article. Review.
-
Melatonin potentiates the antitumor effect of curcumin by inhibiting IKKβ/NF-κB/COX-2 signaling pathway.Int J Oncol. 2017 Oct;51(4):1249-1260. doi: 10.3892/ijo.2017.4097. Epub 2017 Aug 22. Int J Oncol. 2017. PMID: 28849163 Free PMC article.
-
MYC Oncogene: A Druggable Target for Treating Cancers with Natural Products.Aging Dis. 2024 Apr 1;15(2):640-697. doi: 10.14336/AD.2023.0520. Aging Dis. 2024. PMID: 37450923 Free PMC article. Review.
-
Combination Therapy with a TLR7 Agonist and a BRD4 Inhibitor Suppresses Tumor Growth via Enhanced Immunomodulation.Int J Mol Sci. 2024 Jan 4;25(1):663. doi: 10.3390/ijms25010663. Int J Mol Sci. 2024. PMID: 38203835 Free PMC article.
References
-
- Fujiwara K, Ghosh S, Liang P, Morien E, Soma M, Nagase H. Genome-wide screening of aberrant DNA methylation which associated with gene expression in mouse skin cancers. Molecular carcinogenesis. 2013 - PubMed
-
- Gambichler T, Sand M, Skrygan M. Loss of 5-hydroxymethylcytosine and ten-eleven translocation 2 protein expression in malignant melanoma. Melanoma research. 2013;23(3):218–20. - PubMed
-
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, Bourque C, Burke CJ, Turner L, Uong A, Johnson LA, Beroukhim R, Mermel CH, Loda M, Ait-Si-Ali S, Garraway LA, Young RA, Zon LI. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471(7339):513–7. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

