Multiple Independent Retroelement Insertions in the Promoter of a Stress Response Gene Have Variable Molecular and Functional Effects in Drosophila
- PMID: 27517860
- PMCID: PMC4982627
- DOI: 10.1371/journal.pgen.1006249
Multiple Independent Retroelement Insertions in the Promoter of a Stress Response Gene Have Variable Molecular and Functional Effects in Drosophila
Abstract
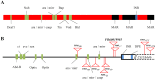
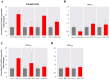
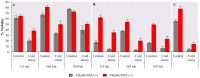
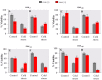
Promoters are structurally and functionally diverse gene regulatory regions. The presence or absence of sequence motifs and the spacing between the motifs defines the properties of promoters. Recent alternative promoter usage analyses in Drosophila melanogaster revealed that transposable elements significantly contribute to promote diversity. In this work, we analyzed in detail one of the transposable element insertions, named FBti0019985, that has been co-opted to drive expression of CG18446, a candidate stress response gene. We analyzed strains from different natural populations and we found that besides FBti0019985, there are another eight independent transposable elements inserted in the proximal promoter region of CG18446. All nine insertions are solo-LTRs that belong to the roo family. We analyzed the sequence of the nine roo insertions and we investigated whether the different insertions were functionally equivalent by performing 5'-RACE, gene expression, and cold-stress survival experiments. We found that different insertions have different molecular and functional consequences. The exact position where the transposable elements are inserted matters, as they all showed highly conserved sequences but only two of the analyzed insertions provided alternative transcription start sites, and only the FBti0019985 insertion consistently affects CG18446 expression. The phenotypic consequences of the different insertions also vary: only FBti0019985 was associated with cold-stress tolerance. Interestingly, the only previous report of transposable elements inserting repeatedly and independently in a promoter region in D. melanogaster, were also located upstream of a stress response gene. Our results suggest that functional validation of individual structural variants is needed to resolve the complexity of insertion clusters.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Contrasting patterns of transposable element insertions in Drosophila heat-shock promoters.PLoS One. 2009 Dec 29;4(12):e8486. doi: 10.1371/journal.pone.0008486. PLoS One. 2009. PMID: 20041194 Free PMC article.
-
A unique cluster of roo insertions in the promoter region of a stress response gene in Drosophila melanogaster.Mob DNA. 2019 Mar 13;10:10. doi: 10.1186/s13100-019-0152-9. eCollection 2019. Mob DNA. 2019. PMID: 30911338 Free PMC article.
-
Regulatory regions in natural transposable element insertions drive interindividual differences in response to immune challenges in Drosophila.Genome Biol. 2021 Sep 14;22(1):265. doi: 10.1186/s13059-021-02471-3. Genome Biol. 2021. PMID: 34521452 Free PMC article.
-
Mobile DNA transposition in somatic cells.BMC Biol. 2011 Sep 29;9:62. doi: 10.1186/1741-7007-9-62. BMC Biol. 2011. PMID: 21958341 Free PMC article. Review.
-
Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster.Bioessays. 1994 Apr;16(4):269-75. doi: 10.1002/bies.950160410. Bioessays. 1994. PMID: 8031304 Review.
Cited by
-
Genome-wide patterns of local adaptation in Western European Drosophila melanogaster natural populations.Sci Rep. 2018 Nov 1;8(1):16143. doi: 10.1038/s41598-018-34267-0. Sci Rep. 2018. PMID: 30385770 Free PMC article.
-
Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila.PLoS Genet. 2019 Feb 12;15(2):e1007900. doi: 10.1371/journal.pgen.1007900. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30753202 Free PMC article.
-
Transposable Elements Contribute to the Adaptation of Arabidopsis thaliana.Genome Biol Evol. 2018 Aug 1;10(8):2140-2150. doi: 10.1093/gbe/evy171. Genome Biol Evol. 2018. PMID: 30102348 Free PMC article.
-
The Interplay Between Developmental Stage and Environment Underlies the Adaptive Effect of a Natural Transposable Element Insertion.Mol Biol Evol. 2023 Mar 4;40(3):msad044. doi: 10.1093/molbev/msad044. Mol Biol Evol. 2023. PMID: 36811953 Free PMC article.
-
Taming, Domestication and Exaptation: Trajectories of Transposable Elements in Genomes.Cells. 2021 Dec 20;10(12):3590. doi: 10.3390/cells10123590. Cells. 2021. PMID: 34944100 Free PMC article. Review.
References
-
- Haberle V, Lenhard B. Promoter architectures and developmental gene regulation. Semin Cell Dev Biol. 2016. - PubMed
-
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38(6):626–35. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

