The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae
- PMID: 27516616
- PMCID: PMC4981263
- DOI: 10.1534/genetics.112.145243
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae
Abstract
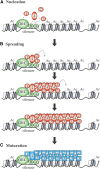
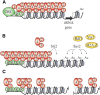
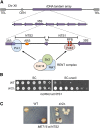
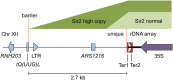
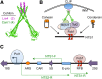
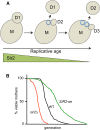
Transcriptional silencing in Saccharomyces cerevisiae occurs at several genomic sites including the silent mating-type loci, telomeres, and the ribosomal DNA (rDNA) tandem array. Epigenetic silencing at each of these domains is characterized by the absence of nearly all histone modifications, including most prominently the lack of histone H4 lysine 16 acetylation. In all cases, silencing requires Sir2, a highly-conserved NAD(+)-dependent histone deacetylase. At locations other than the rDNA, silencing also requires additional Sir proteins, Sir1, Sir3, and Sir4 that together form a repressive heterochromatin-like structure termed silent chromatin. The mechanisms of silent chromatin establishment, maintenance, and inheritance have been investigated extensively over the last 25 years, and these studies have revealed numerous paradigms for transcriptional repression, chromatin organization, and epigenetic gene regulation. Studies of Sir2-dependent silencing at the rDNA have also contributed to understanding the mechanisms for maintaining the stability of repetitive DNA and regulating replicative cell aging. The goal of this comprehensive review is to distill a wide array of biochemical, molecular genetic, cell biological, and genomics studies down to the "nuts and bolts" of silent chromatin and the processes that yield transcriptional silencing.
Keywords: Sir2; chromatin; histone deacetylation; silencing; yeast.
Copyright © 2016 by the Genetics Society of America.
Figures



Similar articles
-
Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation.Mol Cell Biol. 2002 Jun;22(12):4167-80. doi: 10.1128/MCB.22.12.4167-4180.2002. Mol Cell Biol. 2002. PMID: 12024030 Free PMC article.
-
A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors.Mol Cell Biol. 1999 Apr;19(4):3184-97. doi: 10.1128/MCB.19.4.3184. Mol Cell Biol. 1999. PMID: 10082585 Free PMC article.
-
A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin.Mol Cell Biol. 2005 Jun;25(11):4514-28. doi: 10.1128/MCB.25.11.4514-4528.2005. Mol Cell Biol. 2005. PMID: 15899856 Free PMC article.
-
A model for step-wise assembly of heterochromatin in yeast.Novartis Found Symp. 2004;259:48-56; discussion 56-62, 163-9. Novartis Found Symp. 2004. PMID: 15171246 Review.
-
The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more.Curr Opin Microbiol. 2000 Apr;3(2):132-7. doi: 10.1016/s1369-5274(00)00064-3. Curr Opin Microbiol. 2000. PMID: 10744999 Review.
Cited by
-
Gene repression in S. cerevisiae-looking beyond Sir-dependent gene silencing.Curr Genet. 2021 Feb;67(1):3-17. doi: 10.1007/s00294-020-01114-7. Epub 2020 Oct 10. Curr Genet. 2021. PMID: 33037902 Review.
-
In vivo chromatin organization on native yeast telomeric regions is independent of a cis-telomere loopback conformation.Epigenetics Chromatin. 2020 May 22;13(1):23. doi: 10.1186/s13072-020-00344-w. Epigenetics Chromatin. 2020. PMID: 32443982 Free PMC article.
-
Genome stability is guarded by yeast Rtt105 through multiple mechanisms.Genetics. 2021 Feb 9;217(2):iyaa035. doi: 10.1093/genetics/iyaa035. Genetics. 2021. PMID: 33724421 Free PMC article.
-
Efficient DNA double-strand break formation at single or multiple defined sites in the Saccharomyces cerevisiae genome.Nucleic Acids Res. 2020 Nov 18;48(20):e115. doi: 10.1093/nar/gkaa833. Nucleic Acids Res. 2020. PMID: 33053188 Free PMC article.
-
Cell volume homeostatically controls the rDNA repeat copy number and rRNA synthesis rate in yeast.PLoS Genet. 2021 Apr 7;17(4):e1009520. doi: 10.1371/journal.pgen.1009520. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33826644 Free PMC article.
References
-
- Abraham J., Nasmyth K., Strathern J. N., Klar A. J. S., Hicks J. B., 1984. Regulation of mating-type information in yeast. J. Mol. Biol. 176: 307–331. - PubMed
-
- Ai W., Bertram P. G., Tsang C. K., Chan T.-F., Zheng X. F. S., 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10: 1295–1305. - PubMed
-
- Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., et al. , 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277: 18881–18890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

