Programmed Cell Death During Caenorhabditis elegans Development
- PMID: 27516615
- PMCID: PMC4981262
- DOI: 10.1534/genetics.115.186247
Programmed Cell Death During Caenorhabditis elegans Development
Abstract
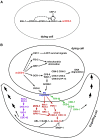
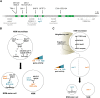
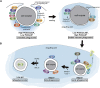
Programmed cell death is an integral component of Caenorhabditis elegans development. Genetic and reverse genetic studies in C. elegans have led to the identification of many genes and conserved cell death pathways that are important for the specification of which cells should live or die, the activation of the suicide program, and the dismantling and removal of dying cells. Molecular, cell biological, and biochemical studies have revealed the underlying mechanisms that control these three phases of programmed cell death. In particular, the interplay of transcriptional regulatory cascades and networks involving multiple transcriptional regulators is crucial in activating the expression of the key death-inducing gene egl-1 and, in some cases, the ced-3 gene in cells destined to die. A protein interaction cascade involving EGL-1, CED-9, CED-4, and CED-3 results in the activation of the key cell death protease CED-3, which is tightly controlled by multiple positive and negative regulators. The activation of the CED-3 caspase then initiates the cell disassembly process by cleaving and activating or inactivating crucial CED-3 substrates; leading to activation of multiple cell death execution events, including nuclear DNA fragmentation, mitochondrial elimination, phosphatidylserine externalization, inactivation of survival signals, and clearance of apoptotic cells. Further studies of programmed cell death in C. elegans will continue to advance our understanding of how programmed cell death is regulated, activated, and executed in general.
Keywords: Caenorhabditis elegans; WormBook; activation phase; execution phase; programmed cell death; specification phase.
Copyright © 2016 by the Genetics Society of America.
Figures

Similar articles
-
Programmed cell death.WormBook. 2005 Oct 6:1-13. doi: 10.1895/wormbook.1.32.1. WormBook. 2005. PMID: 18061982 Free PMC article. Review.
-
Both the caspase CSP-1 and a caspase-independent pathway promote programmed cell death in parallel to the canonical pathway for apoptosis in Caenorhabditis elegans.PLoS Genet. 2013;9(3):e1003341. doi: 10.1371/journal.pgen.1003341. Epub 2013 Mar 7. PLoS Genet. 2013. PMID: 23505386 Free PMC article.
-
DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans.Nature. 2005 Feb 17;433(7027):754-60. doi: 10.1038/nature03316. Nature. 2005. PMID: 15716954
-
Noncanonical cell death in the nematode Caenorhabditis elegans.Methods Enzymol. 2014;545:157-80. doi: 10.1016/B978-0-12-801430-1.00007-X. Methods Enzymol. 2014. PMID: 25065890 Free PMC article. Review.
-
The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9.Cell. 1998 May 15;93(4):519-29. doi: 10.1016/s0092-8674(00)81182-4. Cell. 1998. PMID: 9604928
Cited by
-
Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.Cell Death Discov. 2022 Nov 3;8(1):441. doi: 10.1038/s41420-022-01233-9. Cell Death Discov. 2022. PMID: 36323675 Free PMC article.
-
Loss of the Major Phosphatidylserine or Phosphatidylethanolamine Flippases Differentially Affect Phagocytosis.Front Cell Dev Biol. 2020 Jul 21;8:648. doi: 10.3389/fcell.2020.00648. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32793595 Free PMC article.
-
Ferroptosis regulation by Cap'n'collar family transcription factors.J Biol Chem. 2024 Aug;300(8):107583. doi: 10.1016/j.jbc.2024.107583. Epub 2024 Jul 16. J Biol Chem. 2024. PMID: 39025451 Free PMC article. Review.
-
The nucleoside diphosphate kinase NDK-1/NME1 promotes phagocytosis in concert with DYN-1/Dynamin.FASEB J. 2019 Oct;33(10):11606-11614. doi: 10.1096/fj.201900220R. Epub 2019 Jul 17. FASEB J. 2019. PMID: 31242766 Free PMC article.
-
DRP-1-mediated apoptosis induces muscle degeneration in dystrophin mutants.Sci Rep. 2018 May 9;8(1):7354. doi: 10.1038/s41598-018-25727-8. Sci Rep. 2018. PMID: 29743663 Free PMC article.
References
-
- Abraham M. C., Lu Y., Shaham S., 2007. A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev. Cell 12: 73–86. - PubMed
-
- Adams J. M., 2003. Ways of dying: multiple pathways to apoptosis. Genes Dev. 17: 2481–2495. - PubMed
-
- Adams J. M., Cory S., 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 26: 61–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

