Mammalian PNLDC1 is a novel poly(A) specific exonuclease with discrete expression during early development
- PMID: 27515512
- PMCID: PMC5062988
- DOI: 10.1093/nar/gkw709
Mammalian PNLDC1 is a novel poly(A) specific exonuclease with discrete expression during early development
Abstract
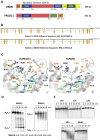
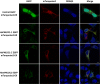
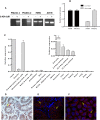
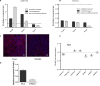
PNLDC1 is a homologue of poly(A) specific ribonuclease (PARN), a known deadenylase with additional role in processing of non-coding RNAs. Both enzymes were reported recently to participate in piRNA biogenesis in silkworm and C. elegans, respectively. To get insights on the role of mammalian PNLDC1, we characterized the human and mouse enzymes. PNLDC1 shows limited conservation compared to PARN and represents an evolutionary related but distinct group of enzymes. It is expressed specifically in mouse embryonic stem cells, human and mouse testes and during early mouse embryo development, while it fades during differentiation. Its expression in differentiated cells, is suppressed through methylation of its promoter by the de novo methyltransferase DNMT3B. Both enzymes are localized mainly in the ER and exhibit in vitro specificity restricted solely to 3' RNA or DNA polyadenylates. Knockdown of Pnldc1 in mESCs and subsequent NGS analysis showed that although the expression of the remaining deadenylases remains unaffected, it affects genes involved mainly in reprogramming, cell cycle and translational regulation. Mammalian PNLDC1 is a novel deadenylase expressed specifically in cell types which share regulatory mechanisms required for multipotency maintenance. Moreover, it could be involved both in posttranscriptional regulation through deadenylation and genome surveillance during early development.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Expanding the repertoire of deadenylases.RNA Biol. 2017 Oct 3;14(10):1320-1325. doi: 10.1080/15476286.2017.1300222. Epub 2017 Mar 7. RNA Biol. 2017. PMID: 28267419 Free PMC article. Review.
-
PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development.EMBO Rep. 2018 Mar;19(3):e44957. doi: 10.15252/embr.201744957. Epub 2018 Feb 14. EMBO Rep. 2018. PMID: 29444933 Free PMC article.
-
Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase.Crit Rev Biochem Mol Biol. 2013 Mar-Apr;48(2):192-209. doi: 10.3109/10409238.2013.771132. Epub 2013 Mar 15. Crit Rev Biochem Mol Biol. 2013. PMID: 23496118 Review.
-
Modulation of poly(A)-specific ribonuclease (PARN): current knowledge and perspectives.Curr Med Chem. 2012;19(28):4838-49. doi: 10.2174/092986712803341539. Curr Med Chem. 2012. PMID: 22834816 Review.
-
Deadenylation: enzymes, regulation, and functional implications.Wiley Interdiscip Rev RNA. 2014 May-Jun;5(3):421-43. doi: 10.1002/wrna.1221. Epub 2014 Feb 12. Wiley Interdiscip Rev RNA. 2014. PMID: 24523229 Review.
Cited by
-
An examination of mediation by DNA methylation on birthweight differences induced by assisted reproductive technologies.Clin Epigenetics. 2022 Nov 28;14(1):151. doi: 10.1186/s13148-022-01381-w. Clin Epigenetics. 2022. PMID: 36443807 Free PMC article.
-
PNLDC1 is essential for piRNA 3' end trimming and transposon silencing during spermatogenesis in mice.Nat Commun. 2017 Oct 10;8(1):819. doi: 10.1038/s41467-017-00854-4. Nat Commun. 2017. PMID: 29018194 Free PMC article.
-
The Function and Prognostic Value of RNA-Binding Proteins in Colorectal Adenocarcinoma Were Analyzed Based on Bioinformatics of Smart Medical Big Data.J Healthc Eng. 2021 Jun 2;2021:5536330. doi: 10.1155/2021/5536330. eCollection 2021. J Healthc Eng. 2021. Retraction in: J Healthc Eng. 2023 May 24;2023:9896539. doi: 10.1155/2023/9896539. PMID: 34188789 Free PMC article. Retracted.
-
Regulatory roles of vertebrate Nocturnin: insights and remaining mysteries.RNA Biol. 2018;15(10):1255-1267. doi: 10.1080/15476286.2018.1526541. Epub 2018 Oct 9. RNA Biol. 2018. PMID: 30257600 Free PMC article.
-
Stage-dependent piRNAs in chicken implicated roles in modulating male germ cell development.BMC Genomics. 2018 Jun 1;19(1):425. doi: 10.1186/s12864-018-4820-9. BMC Genomics. 2018. PMID: 29859049 Free PMC article.
References
-
- Goldstrohm A.C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. - PubMed
-
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. - PubMed
-
- Mitchell P., Tollervey D. mRNA turnover. Curr. Opin. Chem. Biol. 2001;13:320–325. - PubMed
-
- Yan Y.B. Deadenylation: enzymes, regulation, and functional implications. Wiley Interdiscip. Rev. RNA. 2014;5:421–443. - PubMed
-
- Harnisch C., Moritz B., Rammelt C., Temme C., Wahle E. Activity and Function of Deadenylases. Enzymes. 2012;31:181–211. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

