Insights into the Conformation of the Membrane Proximal Regions Critical to the Trimerization of the HIV-1 gp41 Ectodomain Bound to Dodecyl Phosphocholine Micelles
- PMID: 27513582
- PMCID: PMC4981318
- DOI: 10.1371/journal.pone.0160597
Insights into the Conformation of the Membrane Proximal Regions Critical to the Trimerization of the HIV-1 gp41 Ectodomain Bound to Dodecyl Phosphocholine Micelles
Abstract
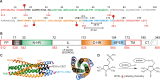
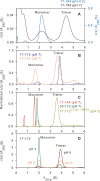
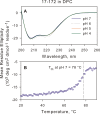
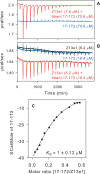
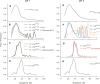
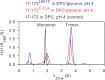
The transitioning of the ectodomain of gp41 from a pre-hairpin to a six-helix bundle conformation is a crucial aspect of virus-cell fusion. To gain insight into the intermediary steps of the fusion process we have studied the pH and dodecyl phosphocholine (DPC) micelle dependent trimer association of gp41 by systematic deletion analysis of an optimized construct termed 17-172 (residues 528 to 683 of Env) that spans the fusion peptide proximal region (FPPR) to the membrane proximal external region (MPER) of gp41, by sedimentation velocity and double electron-electron resonance (DEER) EPR spectroscopy. Trimerization at pH 7 requires the presence of both the FPPR and MPER regions. However, at pH 4, the protein completely dissociates to monomers. DEER measurements reveal a partial fraying of the C-terminal MPER residues in the 17-172 trimer while the other regions, including the FPPR, remain compact. In accordance, truncating nine C-terminal MPER residues (675-683) in the 17-172 construct does not shift the trimer-monomer equilibrium significantly. Thus, in the context of the gp41 ectodomain spanning residues 17-172, trimerization is clearly dependent on FPPR and MPER regions even when the terminal residues of MPER unravel. The antibody Z13e1, which spans both the 2F5 and 4E10 epitopes in MPER, binds to 17-172 with a Kd of 1 ± 0.12 μM. Accordingly, individual antibodies 2F5 and 4E10 also recognize the 17-172 trimer/DPC complex. We propose that binding of the C-terminal residues of MPER to the surface of the DPC micelles models a correct positioning of the trimeric transmembrane domain anchored in the viral membrane.
Conflict of interest statement
Figures
Similar articles
-
Conditional trimerization and lytic activity of HIV-1 gp41 variants containing the membrane-associated segments.Biochemistry. 2015 Mar 3;54(8):1589-99. doi: 10.1021/bi501376f. Epub 2015 Feb 13. Biochemistry. 2015. PMID: 25658332 Free PMC article.
-
An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10.J Virol. 2007 Apr;81(8):4033-43. doi: 10.1128/JVI.02588-06. Epub 2007 Feb 7. J Virol. 2007. PMID: 17287272 Free PMC article.
-
Fully hydrophobic HIV gp41 adopts a hemifusion-like conformation in phospholipid bilayers.J Biol Chem. 2019 Oct 4;294(40):14732-14744. doi: 10.1074/jbc.RA119.009542. Epub 2019 Aug 13. J Biol Chem. 2019. PMID: 31409642 Free PMC article.
-
Antigp41 membrane proximal external region antibodies and the art of using the membrane for neutralization.Curr Opin HIV AIDS. 2017 May;12(3):250-256. doi: 10.1097/COH.0000000000000364. Curr Opin HIV AIDS. 2017. PMID: 28422789 Review.
-
Liposome-based peptide vaccines to elicit immune responses against the membrane active domains of the HIV-1 Env glycoprotein.Biochim Biophys Acta Biomembr. 2024 Jan;1866(1):184235. doi: 10.1016/j.bbamem.2023.184235. Epub 2023 Oct 2. Biochim Biophys Acta Biomembr. 2024. PMID: 37793559 Review.
Cited by
-
Conformational and lipid bilayer-perturbing properties of Marburg virus GP2 segments containing the fusion loop and membrane-proximal external region/transmembrane domain.Heliyon. 2019 Dec 12;5(12):e03018. doi: 10.1016/j.heliyon.2019.e03018. eCollection 2019 Dec. Heliyon. 2019. PMID: 31890962 Free PMC article.
-
Efficient Fusion at Neutral pH by Human Immunodeficiency Virus gp41 Trimers Containing the Fusion Peptide and Transmembrane Domains.Biochemistry. 2018 Feb 20;57(7):1219-1235. doi: 10.1021/acs.biochem.7b00753. Epub 2018 Feb 6. Biochemistry. 2018. PMID: 29345922 Free PMC article.
-
Transmembrane Helix Integrity versus Fraying To Expose Hydrogen Bonds at a Membrane-Water Interface.Biochemistry. 2019 Feb 12;58(6):633-645. doi: 10.1021/acs.biochem.8b01119. Epub 2019 Jan 3. Biochemistry. 2019. PMID: 30565458 Free PMC article.
-
Structure of HIV-1 gp41 with its membrane anchors targeted by neutralizing antibodies.Elife. 2021 Apr 19;10:e65005. doi: 10.7554/eLife.65005. Elife. 2021. PMID: 33871352 Free PMC article.
-
HIV Transmembrane Glycoprotein Conserved Domains and Genetic Markers Across HIV-1 and HIV-2 Variants.Front Microbiol. 2022 May 27;13:855232. doi: 10.3389/fmicb.2022.855232. eCollection 2022. Front Microbiol. 2022. PMID: 35694284 Free PMC article.
References
-
- Roux KH, Taylor KA (2007) AIDS virus envelope spike structure. Curr Opin Struct Biol 17:244–252. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

