Graf1 Controls the Growth of Human Parainfluenza Virus Type 2 through Inactivation of RhoA Signaling
- PMID: 27512058
- PMCID: PMC5044827
- DOI: 10.1128/JVI.01471-16
Graf1 Controls the Growth of Human Parainfluenza Virus Type 2 through Inactivation of RhoA Signaling
Abstract
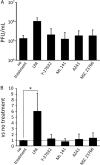
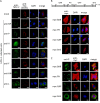
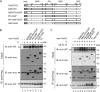
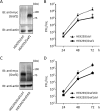
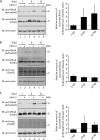
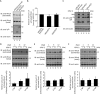
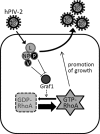
Rho GTPases are involved in a variety of cellular activities and are regulated by guanine nucleotide exchange factors and GTPase-activating proteins (GAPs). We found that the activation of Rho GTPases by lysophosphatidic acid promotes the growth of human parainfluenza virus type 2 (hPIV-2). Furthermore, hPIV-2 infection causes activation of RhoA, a Rho GTPase. We hypothesized that Graf1 (also known as ARHGAP26), a GAP, regulates hPIV-2 growth by controlling RhoA signaling. Immunofluorescence analysis showed that hPIV-2 infection altered Graf1 localization from a homogenous distribution within the cytoplasm to granules. Graf1 colocalized with hPIV-2 P, NP, and L proteins. Graf1 interacts with P and V proteins via their N-terminal common region, and the C-terminal Src homology 3 domain-containing region of Graf1 is important for these interactions. In HEK293 cells constitutively expressing Graf1, hPIV-2 growth was inhibited, and RhoA activation was not observed during hPIV-2 infection. In contrast, Graf1 knockdown restored hPIV-2 growth and RhoA activation. Overexpression of hPIV-2 P and V proteins enhanced hPIV-2-induced RhoA activation. These results collectively suggested that hPIV-2 P and V proteins enhanced hPIV-2 growth by binding to Graf1 and that Graf1 inhibits hPIV-2 growth through RhoA inactivation.
Importance: Robust growth of hPIV-2 requires Rho activation. hPIV-2 infection causes RhoA activation, which is suppressed by Graf1. Graf1 colocalizes with viral RNP (vRNP) in hPIV-2-infected cells. We found that Graf1 interacts with hPIV-2 P and V proteins. We also identified regions in these proteins which are important for this interaction. hPIV-2 P and V proteins enhanced the hPIV-2 growth via binding to Graf1, while Graf1 inhibited hPIV-2 growth through RhoA inactivation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
The V protein of human parainfluenza virus type 2 promotes RhoA-induced filamentous actin formation.Virology. 2018 Nov;524:90-96. doi: 10.1016/j.virol.2018.08.015. Epub 2018 Aug 27. Virology. 2018. PMID: 30165310
-
Common and unique mechanisms of filamentous actin formation by viruses of the genus Orthorubulavirus.Arch Virol. 2020 Apr;165(4):799-807. doi: 10.1007/s00705-020-04565-y. Epub 2020 Feb 25. Arch Virol. 2020. PMID: 32100137
-
Skeletal muscle differentiation and fusion are regulated by the BAR-containing Rho-GTPase-activating protein (Rho-GAP), GRAF1.J Biol Chem. 2011 Jul 22;286(29):25903-21. doi: 10.1074/jbc.M111.243030. Epub 2011 May 26. J Biol Chem. 2011. PMID: 21622574 Free PMC article.
-
The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling.Cell Signal. 2014 Mar;26(3):483-91. doi: 10.1016/j.cellsig.2013.11.034. Epub 2013 Dec 2. Cell Signal. 2014. PMID: 24308970 Review.
-
Mechanisms for spatiotemporal regulation of Rho-GTPase signaling at synapses.Neurosci Lett. 2015 Aug 5;601:4-10. doi: 10.1016/j.neulet.2015.05.034. Epub 2015 May 21. Neurosci Lett. 2015. PMID: 26003445 Free PMC article. Review.
Cited by
-
The role of GTPase-activating protein ARHGAP26 in human cancers.Mol Cell Biochem. 2022 Jan;477(1):319-326. doi: 10.1007/s11010-021-04274-3. Epub 2021 Oct 30. Mol Cell Biochem. 2022. PMID: 34716859 Free PMC article. Review.
-
Human parainfluenza virus type 2 V protein inhibits induction of tetherin.Med Microbiol Immunol. 2017 Aug;206(4):311-318. doi: 10.1007/s00430-017-0508-z. Epub 2017 Apr 28. Med Microbiol Immunol. 2017. PMID: 28455649
-
Inhibition of Cavin3 Degradation by the Human Parainfluenza Virus Type 2 V Protein Is Important for Efficient Viral Growth.Front Microbiol. 2020 Apr 30;11:803. doi: 10.3389/fmicb.2020.00803. eCollection 2020. Front Microbiol. 2020. PMID: 32425917 Free PMC article.
-
Human Parainfluenza Virus Type 2 V Protein Modulates Iron Homeostasis.J Virol. 2021 Feb 24;95(6):e01861-20. doi: 10.1128/JVI.01861-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33408172 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

