Influenza A Virus Infection in Pigs Attracts Multifunctional and Cross-Reactive T Cells to the Lung
- PMID: 27512056
- PMCID: PMC5044846
- DOI: 10.1128/JVI.01211-16
Influenza A Virus Infection in Pigs Attracts Multifunctional and Cross-Reactive T Cells to the Lung
Abstract
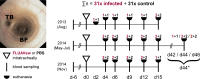
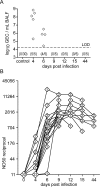
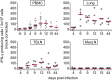
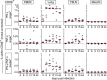
Pigs are natural hosts for influenza A viruses and play a critical role in influenza epidemiology. However, little is known about their influenza-evoked T-cell response. We performed a thorough analysis of both the local and systemic T-cell response in influenza virus-infected pigs, addressing kinetics and phenotype as well as multifunctionality (gamma interferon [IFN-γ], tumor necrosis factor alpha [TNF-α], and interleukin-2 [IL-2]) and cross-reactivity. A total of 31 pigs were intratracheally infected with an H1N2 swine influenza A virus (FLUAVsw) and consecutively euthanized. Lungs, tracheobronchial lymph nodes, and blood were sampled during the first 15 days postinfection (p.i.) and at 6 weeks p.i. Ex vivo flow cytometry of lung lymphocytes revealed an increase in proliferating (Ki-67(+)) CD8(+) T cells with an early effector phenotype (perforin(+) CD27(+)) at day 6 p.i. Low frequencies of influenza virus-specific IFN-γ-producing CD4(+) and CD8(+) T cells could be detected in the lung as early as 4 days p.i. On consecutive days, influenza virus-specific CD4(+) and CD8(+) T cells produced mainly IFN-γ and/or TNF-α, reaching peak frequencies around day 9 p.i., which were up to 30-fold higher in the lung than in tracheobronchial lymph nodes or blood. At 6 weeks p.i., CD4(+) and CD8(+) memory T cells had accumulated in lung tissue. These cells showed diverse cytokine profiles and in vitro reactivity against heterologous influenza virus strains, all of which supports their potential to combat heterologous influenza virus infections in pigs.
Importance: Pigs not only are a suitable large-animal model for human influenza virus infection and vaccine development but also play a central role in the emergence of new pandemic strains. Although promising candidate universal vaccines are tested in pigs and local T cells are the major correlate of heterologous control, detailed and targeted analyses of T-cell responses at the site of infection are scarce. With the present study, we provide the first detailed characterization of magnitude, kinetics, and phenotype of specific T cells recruited to the lungs of influenza virus-infected pigs, and we could demonstrate multifunctionality, cross-reactivity, and memory formation of these cells. This, and ensuing work in the pig, will strengthen the position of this species as a large-animal model for human influenza virus infection and will immediately benefit vaccine development for improved control of influenza virus infections in pigs.
Copyright © 2016 Talker et al.
Figures








Similar articles
-
Magnitude and kinetics of multifunctional CD4+ and CD8β+ T cells in pigs infected with swine influenza A virus.Vet Res. 2015 May 14;46(1):52. doi: 10.1186/s13567-015-0182-3. Vet Res. 2015. PMID: 25971313 Free PMC article.
-
Polyanhydride nanovaccine against swine influenza virus in pigs.Vaccine. 2017 Feb 22;35(8):1124-1131. doi: 10.1016/j.vaccine.2017.01.019. Epub 2017 Jan 20. Vaccine. 2017. PMID: 28117173
-
Aerosol Delivery of a Candidate Universal Influenza Vaccine Reduces Viral Load in Pigs Challenged with Pandemic H1N1 Virus.J Immunol. 2016 Jun 15;196(12):5014-23. doi: 10.4049/jimmunol.1502632. Epub 2016 May 6. J Immunol. 2016. PMID: 27183611 Free PMC article.
-
Porcine innate and adaptative immune responses to influenza and coronavirus infections.Ann N Y Acad Sci. 2006 Oct;1081(1):130-6. doi: 10.1196/annals.1373.014. Ann N Y Acad Sci. 2006. PMID: 17135502 Free PMC article. Review.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
Cited by
-
Identification of a Newly Conserved SLA-II Epitope in a Structural Protein of Swine Influenza Virus.Front Immunol. 2020 Sep 16;11:2083. doi: 10.3389/fimmu.2020.02083. eCollection 2020. Front Immunol. 2020. PMID: 33042120 Free PMC article.
-
T and B Cell Immune Responses to Influenza Viruses in Pigs.Front Immunol. 2019 Feb 5;10:98. doi: 10.3389/fimmu.2019.00098. eCollection 2019. Front Immunol. 2019. PMID: 30804933 Free PMC article. Review.
-
Experimental H1N1pdm09 infection in pigs mimics human seasonal influenza infections.PLoS One. 2019 Sep 20;14(9):e0222943. doi: 10.1371/journal.pone.0222943. eCollection 2019. PLoS One. 2019. PMID: 31539406 Free PMC article.
-
Characterization of the Efficacy of a Split Swine Influenza A Virus Nasal Vaccine Formulated with a Nanoparticle/STING Agonist Combination Adjuvant in Conventional Pigs.Vaccines (Basel). 2023 Nov 10;11(11):1707. doi: 10.3390/vaccines11111707. Vaccines (Basel). 2023. PMID: 38006039 Free PMC article.
-
Pulmonary IFN-γ Causes Lymphocytic Inflammation and Cough Hypersensitivity by Increasing the Number of IFN-γ-Secreting T Lymphocytes.Allergy Asthma Immunol Res. 2022 Nov;14(6):653-673. doi: 10.4168/aair.2022.14.6.653. Allergy Asthma Immunol Res. 2022. PMID: 36426396 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

