Antigenic Fingerprinting of Antibody Response in Humans following Exposure to Highly Pathogenic H7N7 Avian Influenza Virus: Evidence for Anti-PA-X Antibodies
- PMID: 27512055
- PMCID: PMC5044853
- DOI: 10.1128/JVI.01408-16
Antigenic Fingerprinting of Antibody Response in Humans following Exposure to Highly Pathogenic H7N7 Avian Influenza Virus: Evidence for Anti-PA-X Antibodies
Abstract
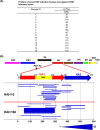
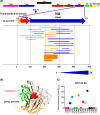
Infections with H7 highly pathogenic avian influenza (HPAI) viruses remain a major public health concern. Adaptation of low-pathogenic H7N7 to highly pathogenic H7N7 in Europe in 2015 raised further alarm for a potential pandemic. An in-depth understanding of antibody responses to HPAI H7 virus following infection in humans could provide important insight into virus gene expression as well as define key protective and serodiagnostic targets. Here we used whole-genome gene fragment phage display libraries (GFPDLs) expressing peptides of 15 to 350 amino acids across the complete genome of the HPAI H7N7 A/Netherlands/33/03 virus. The hemagglutinin (HA) antibody epitope repertoires of 15 H7N7-exposed humans identified clear differences between individuals with no hemagglutination inhibition (HI) titers (<1:10) and those with HI titers of >1:40. Several potentially protective H7N7 epitopes close to the HA receptor binding domain (RBD) and neuraminidase (NA) catalytic site were identified. Surface plasmon resonance (SPR) analysis identified a strong correlation between HA1 (but not HA2) binding antibodies and H7N7 HI titers. A proportion of HA1 binding in plasma was contributed by IgA antibodies. Antibodies against the N7 neuraminidase were less frequent but targeted sites close to the sialic acid binding site. Importantly, we identified strong antibody reactivity against PA-X, a putative virulence factor, in most H7N7-exposed individuals, providing the first evidence for in vivo expression of PA-X and its recognition by the immune system during human influenza A virus infection. This knowledge can help inform the development and selection of the most effective countermeasures for prophylactic as well as therapeutic treatments of HPAI H7N7 avian influenza virus.
Importance: An outbreak of pathogenic H7N7 virus occurred in poultry farms in The Netherlands in 2003. Severe outcome included conjunctivitis, influenza-like illness, and one lethal infection. In this study, we investigated convalescent-phase sera from H7N7-exposed individuals by using a whole-genome phage display library (H7N7-GFPDL) to explore the complete repertoire of post-H7N7-exposure antibodies. PA-X is a recently identified influenza virus virulence protein generated by ribosomal frameshifting in segment 3 of influenza virus coding for PA. However, PA-X expression during influenza virus infection in humans is unknown. We identified strong antibody reactivity against PA-X in most H7N7-exposed individuals (but not in unexposed adults), providing the first evidence for in vivo expression of PA-X and its recognition by the immune system during human infection with pathogenic H7N7 avian influenza virus.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Inactivated H7 Influenza Virus Vaccines Protect Mice despite Inducing Only Low Levels of Neutralizing Antibodies.J Virol. 2017 Sep 27;91(20):e01202-17. doi: 10.1128/JVI.01202-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28768855 Free PMC article.
-
Serum strain-specific or cross-reactive neuraminidase inhibiting antibodies against pandemic А/California/07/2009(H1N1) influenza in healthy volunteers.BMC Res Notes. 2015 Apr 10;8:136. doi: 10.1186/s13104-015-1086-z. BMC Res Notes. 2015. PMID: 25889924 Free PMC article.
-
Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic avian H5N1 influenza virus.Sci Transl Med. 2010 Jan 20;2(15):15ra5. doi: 10.1126/scitranslmed.3000624. Sci Transl Med. 2010. PMID: 20371470
-
The antigenic architecture of the hemagglutinin of influenza H5N1 viruses.Mol Immunol. 2013 Dec;56(4):705-19. doi: 10.1016/j.molimm.2013.07.010. Epub 2013 Aug 7. Mol Immunol. 2013. PMID: 23933511 Review.
-
Epitopes in the HA and NA of H5 and H7 avian influenza viruses that are important for antigenic drift.FEMS Microbiol Rev. 2024 May 8;48(3):fuae014. doi: 10.1093/femsre/fuae014. FEMS Microbiol Rev. 2024. PMID: 38734891 Free PMC article. Review.
Cited by
-
Back to the Future for Influenza Preimmunity-Looking Back at Influenza Virus History to Infer the Outcome of Future Infections.Viruses. 2019 Jan 30;11(2):122. doi: 10.3390/v11020122. Viruses. 2019. PMID: 30704019 Free PMC article. Review.
-
Immunosensor-based label-free and multiplex detection of influenza viruses: State of the art.Biosens Bioelectron. 2019 Sep 15;141:111476. doi: 10.1016/j.bios.2019.111476. Epub 2019 Jun 25. Biosens Bioelectron. 2019. PMID: 31272058 Free PMC article. Review.
-
A Potent Germline-like Human Monoclonal Antibody Targets a pH-Sensitive Epitope on H7N9 Influenza Hemagglutinin.Cell Host Microbe. 2017 Oct 11;22(4):471-483.e5. doi: 10.1016/j.chom.2017.08.011. Epub 2017 Sep 28. Cell Host Microbe. 2017. PMID: 28966056 Free PMC article.
-
AIV polyantigen epitope expressed by recombinant baculovirus induces a systemic immune response in chicken and mouse models.Virol J. 2020 Aug 5;17(1):121. doi: 10.1186/s12985-020-01388-w. Virol J. 2020. PMID: 32758272 Free PMC article.
-
RNA structure interactions and ribonucleoprotein processes of the influenza A virus.Brief Funct Genomics. 2018 Nov 26;17(6):402-414. doi: 10.1093/bfgp/elx028. Brief Funct Genomics. 2018. PMID: 29040388 Free PMC article. Review.
References
-
- Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in The Netherlands. Lancet 363:587–593. doi:10.1016/S0140-6736(04)15589-X. - DOI - PubMed
-
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 101:1356–1361. doi:10.1073/pnas.0308352100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

