Regulation of energy homeostasis by the ubiquitin-independent REGγ proteasome
- PMID: 27511885
- PMCID: PMC4987533
- DOI: 10.1038/ncomms12497
Regulation of energy homeostasis by the ubiquitin-independent REGγ proteasome
Abstract
Maintenance of energy homeostasis is essential for cell survival. Here, we report that the ATP- and ubiquitin-independent REGγ-proteasome system plays a role in maintaining energy homeostasis and cell survival during energy starvation via repressing rDNA transcription, a major intracellular energy-consuming process. Mechanistically, REGγ-proteasome limits cellular rDNA transcription and energy consumption by targeting the rDNA transcription activator SirT7 for ubiquitin-independent degradation under normal conditions. Moreover, energy starvation induces an AMPK-directed SirT7 phosphorylation and subsequent REGγ-dependent SirT7 subcellular redistribution and degradation, thereby further reducing rDNA transcription to save energy to overcome cell death. Energy starvation is a promising strategy for cancer therapy. Our report also shows that REGγ knockdown markedly improves the anti-tumour activity of energy metabolism inhibitors in mice. Our results underscore a control mechanism for an ubiquitin-independent process in maintaining energy homeostasis and cell viability under starvation conditions, suggesting that REGγ-proteasome inhibition has a potential to provide tumour-starving benefits.
Figures


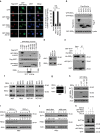
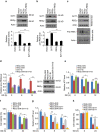
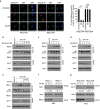
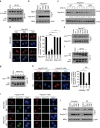

Similar articles
-
The REGγ proteasome regulates hepatic lipid metabolism through inhibition of autophagy.Cell Metab. 2013 Sep 3;18(3):380-91. doi: 10.1016/j.cmet.2013.08.012. Cell Metab. 2013. PMID: 24011073 Free PMC article.
-
Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway.Mol Cell. 2007 Jun 22;26(6):831-42. doi: 10.1016/j.molcel.2007.05.028. Mol Cell. 2007. PMID: 17588518
-
Site-specific acetylation of the proteasome activator REGγ directs its heptameric structure and functions.J Biol Chem. 2013 Jun 7;288(23):16567-16578. doi: 10.1074/jbc.M112.437129. Epub 2013 Apr 23. J Biol Chem. 2013. PMID: 23612972 Free PMC article.
-
Regulation of Life & Death by REGγ.Cells. 2022 Jul 23;11(15):2281. doi: 10.3390/cells11152281. Cells. 2022. PMID: 35892577 Free PMC article. Review.
-
REGgamma, a proteasome activator and beyond?Cell Mol Life Sci. 2008 Dec;65(24):3971-80. doi: 10.1007/s00018-008-8291-z. Cell Mol Life Sci. 2008. PMID: 18679578 Free PMC article. Review.
Cited by
-
O-GlcNAcylation and stablization of SIRT7 promote pancreatic cancer progression by blocking the SIRT7-REGγ interaction.Cell Death Differ. 2022 Oct;29(10):1970-1981. doi: 10.1038/s41418-022-00984-3. Epub 2022 Apr 14. Cell Death Differ. 2022. PMID: 35422493 Free PMC article.
-
The ATM and ATR kinases regulate centrosome clustering and tumor recurrence by targeting KIFC1 phosphorylation.Nat Commun. 2021 Jan 4;12(1):20. doi: 10.1038/s41467-020-20208-x. Nat Commun. 2021. PMID: 33397932 Free PMC article.
-
Red Blood Cell Proteasome in Beta-Thalassemia Trait: Topology of Activity and Networking in Blood Bank Conditions.Membranes (Basel). 2021 Sep 17;11(9):716. doi: 10.3390/membranes11090716. Membranes (Basel). 2021. PMID: 34564533 Free PMC article.
-
Unraveling the Mystery of Energy-Sensing Enzymes and Signaling Pathways in Tumorigenesis and Their Potential as Therapeutic Targets for Cancer.Cells. 2024 Sep 2;13(17):1474. doi: 10.3390/cells13171474. Cells. 2024. PMID: 39273044 Free PMC article. Review.
-
Investigating Physiopathological Roles for Sirtuins in a Mouse Model.Methods Mol Biol. 2023;2589:95-110. doi: 10.1007/978-1-0716-2788-4_7. Methods Mol Biol. 2023. PMID: 36255620
References
-
- Kim K. H. & Lee M. S. Autophagy as a crosstalk mediator of metabolic organs in regulation of energy metabolism. Rev. Endocr. Metab. Dis. 15, 11–20 (2014). - PubMed
-
- Grummt I. & Pikaard C. S. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell. Bio. 4, 641–649 (2003). - PubMed
-
- Murayama A. et al.. Epigenetic Control of rDNA Loci in Response to Intracellular Energy Status. Cell 133, 627–639 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

