Crystallographic snapshots of sulfur insertion by lipoyl synthase
- PMID: 27506792
- PMCID: PMC5003258
- DOI: 10.1073/pnas.1602486113
Crystallographic snapshots of sulfur insertion by lipoyl synthase
Abstract
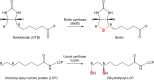
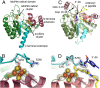
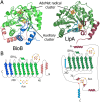
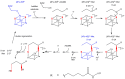
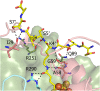
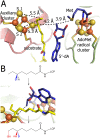
Lipoyl synthase (LipA) catalyzes the insertion of two sulfur atoms at the unactivated C6 and C8 positions of a protein-bound octanoyl chain to produce the lipoyl cofactor. To activate its substrate for sulfur insertion, LipA uses a [4Fe-4S] cluster and S-adenosylmethionine (AdoMet) radical chemistry; the remainder of the reaction mechanism, especially the source of the sulfur, has been less clear. One controversial proposal involves the removal of sulfur from a second (auxiliary) [4Fe-4S] cluster on the enzyme, resulting in destruction of the cluster during each round of catalysis. Here, we present two high-resolution crystal structures of LipA from Mycobacterium tuberculosis: one in its resting state and one at an intermediate state during turnover. In the resting state, an auxiliary [4Fe-4S] cluster has an unusual serine ligation to one of the irons. After reaction with an octanoyllysine-containing 8-mer peptide substrate and 1 eq AdoMet, conditions that allow for the first sulfur insertion but not the second insertion, the serine ligand dissociates from the cluster, the iron ion is lost, and a sulfur atom that is still part of the cluster becomes covalently attached to C6 of the octanoyl substrate. This intermediate structure provides a clear picture of iron-sulfur cluster destruction in action, supporting the role of the auxiliary cluster as the sulfur source in the LipA reaction and describing a radical strategy for sulfur incorporation into completely unactivated substrates.
Keywords: iron–sulfur cluster; lipoic acid; radical SAM enzyme.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The A-type domain in Escherichia coli NfuA is required for regenerating the auxiliary [4Fe-4S] cluster in Escherichia coli lipoyl synthase.J Biol Chem. 2019 Feb 1;294(5):1609-1617. doi: 10.1074/jbc.RA118.006171. Epub 2018 Dec 11. J Biol Chem. 2019. PMID: 30538130 Free PMC article.
-
Trapping a cross-linked lysine-tryptophan radical in the catalytic cycle of the radical SAM enzyme SuiB.Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2101571118. doi: 10.1073/pnas.2101571118. Proc Natl Acad Sci U S A. 2021. PMID: 34001621 Free PMC article.
-
Biochemical Approaches to Probe the Role of the Auxiliary Iron-Sulfur Cluster of Lipoyl Synthase from Mycobacterium Tuberculosis.Methods Mol Biol. 2021;2353:307-332. doi: 10.1007/978-1-0716-1605-5_16. Methods Mol Biol. 2021. PMID: 34292556
-
The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase.Arch Biochem Biophys. 2005 Jan 1;433(1):312-21. doi: 10.1016/j.abb.2004.10.003. Arch Biochem Biophys. 2005. PMID: 15581586 Review.
-
Biotin synthase: insights into radical-mediated carbon-sulfur bond formation.Biochim Biophys Acta. 2012 Nov;1824(11):1213-22. doi: 10.1016/j.bbapap.2012.01.010. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22326745 Review.
Cited by
-
Characterization of LipS1 and LipS2 from Thermococcus kodakarensis: Proteins Annotated as Biotin Synthases, which Together Catalyze Formation of the Lipoyl Cofactor.ACS Bio Med Chem Au. 2022 Oct 19;2(5):509-520. doi: 10.1021/acsbiomedchemau.2c00018. Epub 2022 Jul 14. ACS Bio Med Chem Au. 2022. PMID: 36281299 Free PMC article.
-
Discovery of a Biotin Synthase That Utilizes an Auxiliary 4Fe-5S Cluster for Sulfur Insertion.J Am Chem Soc. 2024 Jan 24;146(3):1860-1873. doi: 10.1021/jacs.3c05481. Epub 2024 Jan 12. J Am Chem Soc. 2024. PMID: 38215281
-
Advances in synthesis of biotin and assembly of lipoic acid.Curr Opin Chem Biol. 2018 Dec;47:60-66. doi: 10.1016/j.cbpa.2018.08.004. Epub 2018 Sep 17. Curr Opin Chem Biol. 2018. PMID: 30236800 Free PMC article. Review.
-
Iron-sulfur protein odyssey: exploring their cluster functional versatility and challenging identification.Metallomics. 2024 May 2;16(5):mfae025. doi: 10.1093/mtomcs/mfae025. Metallomics. 2024. PMID: 38744662 Free PMC article. Review.
-
Insight into the reaction mechanism of lipoyl synthase: a QM/MM study.J Biol Inorg Chem. 2018 Mar;23(2):221-229. doi: 10.1007/s00775-017-1522-8. Epub 2017 Dec 4. J Biol Inorg Chem. 2018. PMID: 29204715 Free PMC article.
References
-
- Miller JR, et al. Escherichia coli LipA is a lipoyl synthase: In vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry. 2000;39(49):15166–15178. - PubMed
-
- White RH. Stable isotope studies on the biosynthesis of lipoic acid in Escherichia coli. Biochemistry. 1980;19(1):15–19. - PubMed
-
- Douglas P, Kriek M, Bryant P, Roach PL. Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew Chem Int Ed Engl. 2006;45(31):5197–5199. - PubMed
-
- Cicchillo RM, et al. Lipoyl synthase requires two equivalents of S-adenosyl-l-methionine to synthesize one equivalent of lipoic acid. Biochemistry. 2004;43(21):6378–6386. - PubMed
-
- Lieder KW, et al. S-Adenosylmethionine-dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron-sulfur centers by electron paramagnetic resonance. Biochemistry. 1998;37(8):2578–2585. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

