Memory of Inflammation in Regulatory T Cells
- PMID: 27499023
- PMCID: PMC4996371
- DOI: 10.1016/j.cell.2016.07.006
Memory of Inflammation in Regulatory T Cells
Abstract
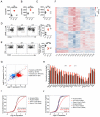
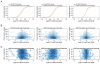
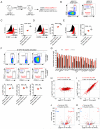
Eukaryotic cells can "remember" transient encounters with a wide range of stimuli, inducing lasting states of altered responsiveness. Regulatory T (Treg) cells are a specialized lineage of suppressive CD4 T cells that act as critical negative regulators of inflammation in various biological contexts. Treg cells exposed to inflammatory conditions acquire strongly enhanced suppressive function. Using inducible genetic tracing, we analyzed the long-term stability of activation-induced transcriptional, epigenomic, and functional changes in Treg cells. We found that the inflammation-experienced Treg cell population reversed many activation-induced changes and lost its enhanced suppressive function over time. The "memory-less" potentiation of Treg suppressor function may help avoid a state of generalized immunosuppression that could otherwise result from repeated activation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells.Immunity. 2012 Feb 24;36(2):262-75. doi: 10.1016/j.immuni.2011.12.012. Epub 2012 Feb 9. Immunity. 2012. PMID: 22326580
-
An NF-κB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function.Immunity. 2017 Sep 19;47(3):450-465.e5. doi: 10.1016/j.immuni.2017.08.010. Epub 2017 Sep 7. Immunity. 2017. PMID: 28889947 Free PMC article.
-
CD27/CFSE-based ex vivo selection of highly suppressive alloantigen-specific human regulatory T cells.J Immunol. 2005 Jun 15;174(12):7573-83. doi: 10.4049/jimmunol.174.12.7573. J Immunol. 2005. PMID: 15944257
-
Lineage stability and phenotypic plasticity of Foxp3⁺ regulatory T cells.Immunol Rev. 2014 May;259(1):159-72. doi: 10.1111/imr.12175. Immunol Rev. 2014. PMID: 24712465 Review.
-
From stability to dynamics: understanding molecular mechanisms of regulatory T cells through Foxp3 transcriptional dynamics.Clin Exp Immunol. 2019 Jul;197(1):14-23. doi: 10.1111/cei.13194. Epub 2018 Sep 17. Clin Exp Immunol. 2019. PMID: 30076771 Free PMC article. Review.
Cited by
-
Sustained store-operated calcium entry utilizing activated chromatin state leads to instability in iTregs.Elife. 2023 Dec 6;12:RP88874. doi: 10.7554/eLife.88874. Elife. 2023. PMID: 38055613 Free PMC article.
-
Induction of anergic or regulatory tumor-specific CD4+ T cells in the tumor-draining lymph node.Nat Commun. 2018 May 29;9(1):2113. doi: 10.1038/s41467-018-04524-x. Nat Commun. 2018. PMID: 29844317 Free PMC article.
-
Type I interferon signaling attenuates regulatory T cell function in viral infection and in the tumor microenvironment.PLoS Pathog. 2018 Apr 19;14(4):e1006985. doi: 10.1371/journal.ppat.1006985. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29672594 Free PMC article.
-
Tbx21 and Foxp3 Are Epigenetically Stabilized in T-Bet+ Tregs That Transiently Accumulate in Influenza A Virus-Infected Lungs.Int J Mol Sci. 2021 Jul 14;22(14):7522. doi: 10.3390/ijms22147522. Int J Mol Sci. 2021. PMID: 34299148 Free PMC article.
-
Emodin modulates gut microbial community and triggers intestinal immunity.J Sci Food Agric. 2023 Feb;103(3):1273-1282. doi: 10.1002/jsfa.12221. Epub 2022 Sep 26. J Sci Food Agric. 2023. PMID: 36088620 Free PMC article.
References
-
- Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annual review of immunology. 2009;27:551–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

