The Exposed Proteomes of Brachyspira hyodysenteriae and B. pilosicoli
- PMID: 27493641
- PMCID: PMC4955376
- DOI: 10.3389/fmicb.2016.01103
The Exposed Proteomes of Brachyspira hyodysenteriae and B. pilosicoli
Abstract
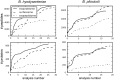
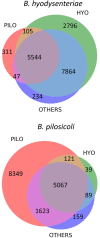
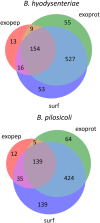
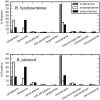
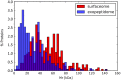
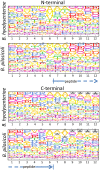
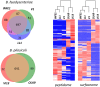
Brachyspira hyodysenteriae and Brachyspira pilosicoli are well-known intestinal pathogens in pigs. B. hyodysenteriae is the causative agent of swine dysentery, a disease with an important impact on pig production while B. pilosicoli is responsible of a milder diarrheal disease in these animals, porcine intestinal spirochetosis. Recent sequencing projects have provided information for the genome of these species facilitating the search of vaccine candidates using reverse vaccinology approaches. However, practically no experimental evidence exists of the actual gene products being expressed and of those proteins exposed on the cell surface or released to the cell media. Using a cell-shaving strategy and a shotgun proteomic approach we carried out a large-scale characterization of the exposed proteins on the bacterial surface in these species as well as of peptides and proteins in the extracellular medium. The study included three strains of B. hyodysenteriae and two strains of B. pilosicoli and involved 148 LC-MS/MS runs on a high resolution Orbitrap instrument. Overall, we provided evidence for more than 29,000 different peptides pointing to 1625 and 1338 different proteins in B. hyodysenteriae and B. pilosicoli, respectively. Many of the most abundant proteins detected corresponded to described virulence factors and vaccine candidates. The level of expression of these proteins, however, was different among species and strains, stressing the value of determining actual gene product levels as a complement of genomic-based approaches for vaccine design.
Keywords: Brachyspira; membrane shaving; shotgun proteomics; surface proteins; virulence factors.
Figures
Similar articles
-
Brachyspira hyodysenteriae and B. pilosicoli Proteins Recognized by Sera of Challenged Pigs.Front Microbiol. 2017 May 4;8:723. doi: 10.3389/fmicb.2017.00723. eCollection 2017. Front Microbiol. 2017. PMID: 28522991 Free PMC article.
-
Analysis of bacterial load and prevalence of mixed infections with Lawsonia intracellularis, Brachyspira hyodysenteriae and/or Brachyspira pilosicoli in German pigs with diarrhoea.Berl Munch Tierarztl Wochenschr. 2011 May-Jun;124(5-6):236-41. Berl Munch Tierarztl Wochenschr. 2011. PMID: 22059295
-
A survey on the occurrence of Brachyspira pilosicoli and Brachyspira hyodysenteriae in growing-finishing pigs.F1000Res. 2019 Sep 30;8:1702. doi: 10.12688/f1000research.20639.3. eCollection 2019. F1000Res. 2019. PMID: 33824718 Free PMC article.
-
The search for Brachyspira outer membrane proteins that interact with the host.Anim Health Res Rev. 2001 Jun;2(1):19-30. Anim Health Res Rev. 2001. PMID: 11708742 Review.
-
A review of methods used for studying the molecular epidemiology of Brachyspira hyodysenteriae.Vet Microbiol. 2017 Aug;207:181-194. doi: 10.1016/j.vetmic.2017.06.011. Epub 2017 Jun 19. Vet Microbiol. 2017. PMID: 28757022 Review.
Cited by
-
Brachyspira hyodysenteriae and B. pilosicoli Proteins Recognized by Sera of Challenged Pigs.Front Microbiol. 2017 May 4;8:723. doi: 10.3389/fmicb.2017.00723. eCollection 2017. Front Microbiol. 2017. PMID: 28522991 Free PMC article.
-
Detection of Serum IgG Specific for Brachyspira pilosicoli and "Brachyspira canis" in Dogs.Vet Sci. 2024 Jul 3;11(7):302. doi: 10.3390/vetsci11070302. Vet Sci. 2024. PMID: 39057986 Free PMC article.
-
The Spirochete Brachyspira pilosicoli, Enteric Pathogen of Animals and Humans.Clin Microbiol Rev. 2017 Nov 29;31(1):e00087-17. doi: 10.1128/CMR.00087-17. Print 2018 Jan. Clin Microbiol Rev. 2017. PMID: 29187397 Free PMC article. Review.
-
Differential expression of hemolysin genes in weakly and strongly hemolytic Brachyspira hyodysenteriae strains.BMC Vet Res. 2020 May 29;16(1):169. doi: 10.1186/s12917-020-02385-5. BMC Vet Res. 2020. PMID: 32471432 Free PMC article.
-
Identification of surface proteins in a clinical Staphylococcus haemolyticus isolate by bacterial surface shaving.BMC Microbiol. 2020 Apr 7;20(1):80. doi: 10.1186/s12866-020-01778-8. BMC Microbiol. 2020. PMID: 32264835 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

