Force and twist dependence of RepC nicking activity on torsionally-constrained DNA molecules
- PMID: 27488190
- PMCID: PMC5062986
- DOI: 10.1093/nar/gkw689
Force and twist dependence of RepC nicking activity on torsionally-constrained DNA molecules
Abstract
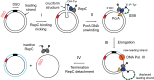
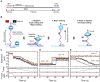
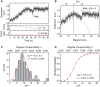
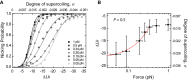
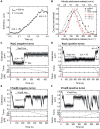
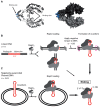
Many bacterial plasmids replicate by an asymmetric rolling-circle mechanism that requires sequence-specific recognition for initiation, nicking of one of the template DNA strands and unwinding of the duplex prior to subsequent leading strand DNA synthesis. Nicking is performed by a replication-initiation protein (Rep) that directly binds to the plasmid double-stranded origin and remains covalently bound to its substrate 5'-end via a phosphotyrosine linkage. It has been proposed that the inverted DNA sequences at the nick site form a cruciform structure that facilitates DNA cleavage. However, the role of Rep proteins in the formation of this cruciform and the implication for its nicking and religation functions is unclear. Here, we have used magnetic tweezers to directly measure the DNA nicking and religation activities of RepC, the replication initiator protein of plasmid pT181, in plasmid sized and torsionally-constrained linear DNA molecules. Nicking by RepC occurred only in negatively supercoiled DNA and was force- and twist-dependent. Comparison with a type IB topoisomerase in similar experiments highlighted a relatively inefficient religation activity of RepC. Based on the structural modeling of RepC and on our experimental evidence, we propose a model where RepC nicking activity is passive and dependent upon the supercoiling degree of the DNA substrate.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Dynamics of DNA nicking and unwinding by the RepC-PcrA complex.Nucleic Acids Res. 2020 Feb 28;48(4):2013-2025. doi: 10.1093/nar/gkz1200. Nucleic Acids Res. 2020. PMID: 31930301 Free PMC article.
-
The PcrA3 mutant binds DNA and interacts with the RepC initiator protein of plasmid pT181 but is defective in its DNA helicase and unwinding activities.Plasmid. 2005 Sep;54(2):104-13. doi: 10.1016/j.plasmid.2005.01.004. Epub 2005 Mar 4. Plasmid. 2005. PMID: 16122559
-
Double-stranded origin nicking and replication initiation are coupled in the replication of a rolling circle plasmid, pT181.FEMS Microbiol Lett. 1997 Jun 15;151(2):185-9. doi: 10.1111/j.1574-6968.1997.tb12568.x. FEMS Microbiol Lett. 1997. PMID: 9228752
-
DNA-protein interactions during the initiation and termination of plasmid pT181 rolling-circle replication.Prog Nucleic Acid Res Mol Biol. 2003;75:113-37. doi: 10.1016/s0079-6603(03)75004-1. Prog Nucleic Acid Res Mol Biol. 2003. PMID: 14604011 Review.
-
Plasmid Rolling-Circle Replication.Microbiol Spectr. 2015 Feb;3(1):PLAS-0035-2014. doi: 10.1128/microbiolspec.PLAS-0035-2014. Microbiol Spectr. 2015. PMID: 26104557 Review.
Cited by
-
Interaction of Proteins with Inverted Repeats and Cruciform Structures in Nucleic Acids.Int J Mol Sci. 2022 May 31;23(11):6171. doi: 10.3390/ijms23116171. Int J Mol Sci. 2022. PMID: 35682854 Free PMC article. Review.
-
Molecular and Functional Characterization of MobK Protein-A Novel-Type Relaxase Involved in Mobilization for Conjugational Transfer of Klebsiella pneumoniae Plasmid pIGRK.Int J Mol Sci. 2021 May 13;22(10):5152. doi: 10.3390/ijms22105152. Int J Mol Sci. 2021. PMID: 34068033 Free PMC article.
-
Dynamics of DNA nicking and unwinding by the RepC-PcrA complex.Nucleic Acids Res. 2020 Feb 28;48(4):2013-2025. doi: 10.1093/nar/gkz1200. Nucleic Acids Res. 2020. PMID: 31930301 Free PMC article.
-
CTP promotes efficient ParB-dependent DNA condensation by facilitating one-dimensional diffusion from parS.Elife. 2021 Jul 12;10:e67554. doi: 10.7554/eLife.67554. Elife. 2021. PMID: 34250901 Free PMC article.
-
Understanding the paradoxical mechanical response of in-phase A-tracts at different force regimes.Nucleic Acids Res. 2020 May 21;48(9):5024-5036. doi: 10.1093/nar/gkaa225. Nucleic Acids Res. 2020. PMID: 32282908 Free PMC article.
References
-
- Ruiz-Maso J.A., Macho N.C., Bordanaba-Ruiseco L., Espinosa M., Coll M., Del Solar G. Plasmid rolling-circle replication. Microbiol. Spectr. 2015;3:1–23. - PubMed
-
- Khan S.A. Mechanism of replication and copy number control of plasmids in gram-positive bacteria. Genet. Eng. (N.Y.) 1996;18:183–201. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

