RNAseq Analyses Identify Tumor Necrosis Factor-Mediated Inflammation as a Major Abnormality in ALS Spinal Cord
- PMID: 27487029
- PMCID: PMC4972368
- DOI: 10.1371/journal.pone.0160520
RNAseq Analyses Identify Tumor Necrosis Factor-Mediated Inflammation as a Major Abnormality in ALS Spinal Cord
Abstract
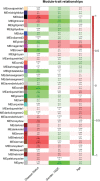
ALS is a rapidly progressive, devastating neurodegenerative illness of adults that produces disabling weakness and spasticity arising from death of lower and upper motor neurons. No meaningful therapies exist to slow ALS progression, and molecular insights into pathogenesis and progression are sorely needed. In that context, we used high-depth, next generation RNA sequencing (RNAseq, Illumina) to define gene network abnormalities in RNA samples depleted of rRNA and isolated from cervical spinal cord sections of 7 ALS and 8 CTL samples. We aligned >50 million 2X150 bp paired-end sequences/sample to the hg19 human genome and applied three different algorithms (Cuffdiff2, DEseq2, EdgeR) for identification of differentially expressed genes (DEG's). Ingenuity Pathways Analysis (IPA) and Weighted Gene Co-expression Network Analysis (WGCNA) identified inflammatory processes as significantly elevated in our ALS samples, with tumor necrosis factor (TNF) found to be a major pathway regulator (IPA) and TNFα-induced protein 2 (TNFAIP2) as a major network "hub" gene (WGCNA). Using the oPOSSUM algorithm, we analyzed transcription factors (TF) controlling expression of the nine DEG/hub genes in the ALS samples and identified TF's involved in inflammation (NFkB, REL, NFkB1) and macrophage function (NR1H2::RXRA heterodimer). Transient expression in human iPSC-derived motor neurons of TNFAIP2 (also a DEG identified by all three algorithms) reduced cell viability and induced caspase 3/7 activation. Using high-density RNAseq, multiple algorithms for DEG identification, and an unsupervised gene co-expression network approach, we identified significant elevation of inflammatory processes in ALS spinal cord with TNF as a major regulatory molecule. Overexpression of the DEG TNFAIP2 in human motor neurons, the population most vulnerable to die in ALS, increased cell death and caspase 3/7 activation. We propose that therapies targeted to reduce inflammatory TNFα signaling may be helpful in ALS patients.
Conflict of interest statement
Figures



Similar articles
-
Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS.Sci Rep. 2017 Aug 30;7(1):10046. doi: 10.1038/s41598-017-10488-7. Sci Rep. 2017. PMID: 28855684 Free PMC article.
-
RNA-seq analyses reveal that cervical spinal cords and anterior motor neurons from amyotrophic lateral sclerosis subjects show reduced expression of mitochondrial DNA-encoded respiratory genes, and rhTFAM may correct this respiratory deficiency.Brain Res. 2017 Jul 15;1667:74-83. doi: 10.1016/j.brainres.2017.05.010. Epub 2017 May 13. Brain Res. 2017. PMID: 28511992
-
Beta-amyloid 42 accumulation in the lumbar spinal cord motor neurons of amyotrophic lateral sclerosis patients.Neurobiol Dis. 2005 Jun-Jul;19(1-2):340-7. doi: 10.1016/j.nbd.2005.01.012. Neurobiol Dis. 2005. PMID: 15837590
-
Spinal cord molecular profiling provides a better understanding of amyotrophic lateral sclerosis pathogenesis.Brain Res Brain Res Rev. 2004 Jul;45(3):213-29. doi: 10.1016/j.brainresrev.2004.04.002. Brain Res Brain Res Rev. 2004. PMID: 15210305 Review.
-
The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients.Brain Res. 2014 Oct 10;1584:28-38. doi: 10.1016/j.brainres.2013.12.011. Epub 2013 Dec 16. Brain Res. 2014. PMID: 24355598 Review.
Cited by
-
Related Endogenous Retrovirus-K Elements Harbor Distinct Protease Active Site Motifs.Front Microbiol. 2018 Jul 18;9:1577. doi: 10.3389/fmicb.2018.01577. eCollection 2018. Front Microbiol. 2018. PMID: 30072963 Free PMC article.
-
ATH-1105, a small-molecule positive modulator of the neurotrophic HGF system, is neuroprotective, preserves neuromotor function, and extends survival in preclinical models of ALS.Front Neurosci. 2024 Feb 8;18:1348157. doi: 10.3389/fnins.2024.1348157. eCollection 2024. Front Neurosci. 2024. PMID: 38389786 Free PMC article.
-
Epigenetic differences between monozygotic twins discordant for amyotrophic lateral sclerosis (ALS) provide clues to disease pathogenesis.PLoS One. 2017 Aug 10;12(8):e0182638. doi: 10.1371/journal.pone.0182638. eCollection 2017. PLoS One. 2017. PMID: 28797086 Free PMC article.
-
Integrated transcriptome landscape of ALS identifies genome instability linked to TDP-43 pathology.Nat Commun. 2023 Apr 20;14(1):2176. doi: 10.1038/s41467-023-37630-6. Nat Commun. 2023. PMID: 37080969 Free PMC article.
-
Nicotinamide Riboside and Pterostilbene Cooperatively Delay Motor Neuron Failure in ALS SOD1G93A Mice.Mol Neurobiol. 2021 Apr;58(4):1345-1371. doi: 10.1007/s12035-020-02188-7. Epub 2020 Nov 10. Mol Neurobiol. 2021. PMID: 33174130
References
-
- Hardiman O, Greenway M. The complex genetics of amyotrophic lateral sclerosis. The LancetNeurology. 2007;6(4):291–2. doi: S1474-4422(07)70062-5 [pii]. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

