Understanding the role of dynamics in the iron sulfur cluster molecular machine
- PMID: 27474202
- PMCID: PMC5176006
- DOI: 10.1016/j.bbagen.2016.07.020
Understanding the role of dynamics in the iron sulfur cluster molecular machine
Abstract
Background: The bacterial proteins IscS, IscU and CyaY, the bacterial orthologue of frataxin, play an essential role in the biological machine that assembles the prosthetic FeS cluster groups on proteins. They form functionally binary and ternary complexes both in vivo and in vitro. Yet, the mechanism by which they work remains unclear.
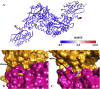
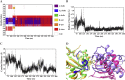
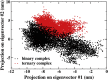
Methods: We carried out extensive molecular dynamics simulations to understand the nature of their interactions and the role of dynamics starting from the crystal structure of a IscS-IscU complex and the experimentally-based model of a ternary IscS-IscU-CyaY complex and used nuclear magnetic resonance to experimentally test the interface.
Results: We show that, while being firmly anchored to IscS, IscU has a pivotal motion around the interface. Our results also describe how the catalytic loop of IscS can flip conformation to allow FeS cluster assembly. This motion is hampered in the ternary complex explaining its inhibitory properties in cluster formation.
Conclusions: We conclude that the observed 'fluid' IscS-IscU interface provides the binary complex with a functional adaptability exploited in partner recognition and unravels the molecular determinants of the reported inhibitory action of CyaY in the IscS-IscU-CyaY complex explained in terms of the hampering effect on specific IscU-IscS movements.
General significance: Our study provides the first mechanistic basis to explain how the IscS-IscU complex selects its binding partners and supports the inhibitory role of CyaY in the ternary complex.
Keywords: CyaY; Frataxin; Iron-sulfur cluster biogenesis; Molecular dynamics; Structure.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
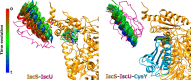




Similar articles
-
Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.PLoS Biol. 2010 Apr 13;8(4):e1000354. doi: 10.1371/journal.pbio.1000354. PLoS Biol. 2010. PMID: 20404999 Free PMC article.
-
The scaffold protein IscU retains a structured conformation in the Fe-S cluster assembly complex.Chembiochem. 2014 Jul 21;15(11):1682-6. doi: 10.1002/cbic.201402211. Epub 2014 Jul 8. Chembiochem. 2014. PMID: 25044349
-
Iron-sulfur cluster biosynthesis: characterization of Escherichia coli CYaY as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold IscU.J Biol Chem. 2006 Jun 16;281(24):16256-63. doi: 10.1074/jbc.M513569200. Epub 2006 Apr 9. J Biol Chem. 2006. PMID: 16603772
-
Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery.FEBS Lett. 2013 Apr 17;587(8):1172-9. doi: 10.1016/j.febslet.2013.01.003. Epub 2013 Jan 16. FEBS Lett. 2013. PMID: 23333622 Free PMC article. Review.
-
Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation.Crit Rev Biochem Mol Biol. 2007 Mar-Apr;42(2):95-111. doi: 10.1080/10409230701322298. Crit Rev Biochem Mol Biol. 2007. PMID: 17453917 Review.
Cited by
-
New Techniques for Ancient Proteins: Direct Coupling Analysis Applied on Proteins Involved in Iron Sulfur Cluster Biogenesis.Front Mol Biosci. 2017 Jun 15;4:40. doi: 10.3389/fmolb.2017.00040. eCollection 2017. Front Mol Biosci. 2017. PMID: 28664160 Free PMC article.
-
A two-hybrid system reveals previously uncharacterized protein-protein interactions within the Helicobacter pylori NIF iron-sulfur maturation system.Sci Rep. 2021 May 24;11(1):10794. doi: 10.1038/s41598-021-90003-1. Sci Rep. 2021. PMID: 34031459 Free PMC article.
-
Cysteine Desulfurase IscS2 Plays a Role in Oxygen Resistance in Clostridium difficile.Infect Immun. 2018 Jul 23;86(8):e00326-18. doi: 10.1128/IAI.00326-18. Print 2018 Aug. Infect Immun. 2018. PMID: 29866903 Free PMC article.
-
Iron-sulfur protein maturation in Helicobacter pylori: identifying a Nfu-type cluster carrier protein and its iron-sulfur protein targets.Mol Microbiol. 2018 May;108(4):379-396. doi: 10.1111/mmi.13942. Epub 2018 Mar 30. Mol Microbiol. 2018. PMID: 29498770 Free PMC article.
-
Hybrid Methods in Iron-Sulfur Cluster Biogenesis.Front Mol Biosci. 2017 Mar 13;4:12. doi: 10.3389/fmolb.2017.00012. eCollection 2017. Front Mol Biosci. 2017. PMID: 28349052 Free PMC article. Review.
References
-
- Roche B., Aussel L., Ezraty B., Mandin P., Py B., Barras F. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta Biomembr. Bioener. 2013;1827:455–469. - PubMed
-
- Roche B., Huguenot A., Barras F., Py B. The iron-binding CyaY and IscX proteins assist the ISC-catalyzed Fe—S biogenesis in Escherichia coli. Mol. Microbiol. 2015;95:605–623. - PubMed
-
- Marelja Z., Stocklein W., Nimtz M., Leimkuhler S. A novel role for human Nfs1 in the cytoplasm - Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J. Biol. Chem. 2008;283:25178–25185. - PubMed
-
- Stehling O., Wilbrecht C., Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

