Short communication: antiviral activity of porcine IFN-λ3 against porcine epidemic diarrhea virus in vitro
- PMID: 27470155
- PMCID: PMC7089062
- DOI: 10.1007/s11262-016-1374-2
Short communication: antiviral activity of porcine IFN-λ3 against porcine epidemic diarrhea virus in vitro
Abstract
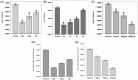
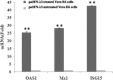
A new family of IFNs called type III IFN or IFN-λ has been described, and shown to induce antiviral activity against several viruses in the cell culture. In this study, the molecular cloning, expression, and antiporcine epidemic diarrhea virus (PEDV) activity of porcine IFN-λ3 (poIFN-λ3) were reported. The full-length poIFN-λ3 cDNA sequence encoded 196 amino acids with a 23 amino acid signal peptide. Sequence alignments showed that poIFN-λ3 had an amino acid sequence similarity to Ovis aries (78.1 %), Bos taurus (76.0 %), Tupaia belangeri (71.3 %), Equus caballus (69.9 %), and Homo sapiens (69.9 %). The phylogenetic analysis based on the genomic sequences indicated that poIFN-λ3 is located in the same branch as B. taurus and O. aries IFN-λ3. The poIFN-λ3 without a signal anchor sequence was efficiently expressed in Escherichia coli, and the purified recombinant poIFN-λ3 exhibited significant antiviral effects against PEDV in a dose- and time-dependent manner. This inhibitory effect of poIFN-λ3 on PEDV was observed under three different treatment conditions. The highest inhibition of PEDV was observed in Vero E6 cell cultures pretreated with poIFN-λ3 (prior to PEDV infection). In addition, poIFN-λ3 was able to induce the expression of IFN-stimulated genes, including ISG15, OAS1, and Mx1 in Vero E6 cells. These data demonstrate that poIFN-λ3 has antiviral activity against PEDV and may serve as a useful biotherapeutic candidate to inhibit PEDV or other viruses in swine.
Keywords: Antiviral activity; PEDV; Porcine IFN-λ3.
Conflict of interest statement
All the authors have no conflict of interest to declare.
Figures


Similar articles
-
Type III Interferon Restriction by Porcine Epidemic Diarrhea Virus and the Role of Viral Protein nsp1 in IRF1 Signaling.J Virol. 2018 Jan 30;92(4):e01677-17. doi: 10.1128/JVI.01677-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187542 Free PMC article.
-
Transcriptomic and antiviral analyses of PoIFN-Delta5 against porcine enteric viruses in porcine intestinal epithelial cells.Vet Microbiol. 2023 May;280:109718. doi: 10.1016/j.vetmic.2023.109718. Epub 2023 Mar 2. Vet Microbiol. 2023. PMID: 36871521
-
IFN-Lambda 3 Mediates Antiviral Protection Against Porcine Epidemic Diarrhea Virus by Inducing a Distinct Antiviral Transcript Profile in Porcine Intestinal Epithelia.Front Immunol. 2019 Oct 17;10:2394. doi: 10.3389/fimmu.2019.02394. eCollection 2019. Front Immunol. 2019. PMID: 31681286 Free PMC article.
-
Coronavirus Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Interacts with p53 To Induce Cell Cycle Arrest in S-Phase and Promotes Viral Replication.J Virol. 2021 Jul 26;95(16):e0018721. doi: 10.1128/JVI.00187-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34037422 Free PMC article.
-
Contribution of molecular biology to the study of the porcine interferon system.Vet Microbiol. 1990 Jun;23(1-4):245-57. doi: 10.1016/0378-1135(90)90155-o. Vet Microbiol. 1990. PMID: 2205970 Free PMC article. Review.
Cited by
-
Manipulation of Intestinal Antiviral Innate Immunity and Immune Evasion Strategies of Porcine Epidemic Diarrhea Virus.Biomed Res Int. 2019 Nov 3;2019:1862531. doi: 10.1155/2019/1862531. eCollection 2019. Biomed Res Int. 2019. PMID: 31781594 Free PMC article. Review.
-
Profile analysis of circRNAs induced by porcine endemic diarrhea virus infection in porcine intestinal epithelial cells.Virology. 2019 Jan 15;527:169-179. doi: 10.1016/j.virol.2018.11.014. Epub 2018 Dec 6. Virology. 2019. PMID: 30530223 Free PMC article.
-
Investigation of the anti-pseudorabies virus activity of interferon lambda 3 in cultured porcine kidney epithelial cells.Vet Med Sci. 2022 Nov;8(6):2444-2450. doi: 10.1002/vms3.933. Epub 2022 Sep 19. Vet Med Sci. 2022. PMID: 36122903 Free PMC article.
-
Type III Interferon Restriction by Porcine Epidemic Diarrhea Virus and the Role of Viral Protein nsp1 in IRF1 Signaling.J Virol. 2018 Jan 30;92(4):e01677-17. doi: 10.1128/JVI.01677-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187542 Free PMC article.
-
The Coronavirus PEDV Evades Type III Interferon Response Through the miR-30c-5p/SOCS1 Axis.Front Microbiol. 2020 May 22;11:1180. doi: 10.3389/fmicb.2020.01180. eCollection 2020. Front Microbiol. 2020. PMID: 32574254 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

