Genome-Wide Prediction and Analysis of 3D-Domain Swapped Proteins in the Human Genome from Sequence Information
- PMID: 27467780
- PMCID: PMC4965083
- DOI: 10.1371/journal.pone.0159627
Genome-Wide Prediction and Analysis of 3D-Domain Swapped Proteins in the Human Genome from Sequence Information
Abstract
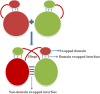
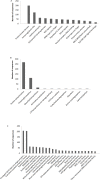
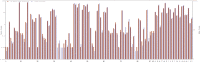
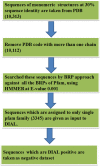
3D-domain swapping is one of the mechanisms of protein oligomerization and the proteins exhibiting this phenomenon have many biological functions. These proteins, which undergo domain swapping, have acquired much attention owing to their involvement in human diseases, such as conformational diseases, amyloidosis, serpinopathies, proteionopathies etc. Early realisation of proteins in the whole human genome that retain tendency to domain swap will enable many aspects of disease control management. Predictive models were developed by using machine learning approaches with an average accuracy of 78% (85.6% of sensitivity, 87.5% of specificity and an MCC value of 0.72) to predict putative domain swapping in protein sequences. These models were applied to many complete genomes with special emphasis on the human genome. Nearly 44% of the protein sequences in the human genome were predicted positive for domain swapping. Enrichment analysis was performed on the positively predicted sequences from human genome for their domain distribution, disease association and functional importance based on Gene Ontology (GO). Enrichment analysis was also performed to infer a better understanding of the functional importance of these sequences. Finally, we developed hinge region prediction, in the given putative domain swapped sequence, by using important physicochemical properties of amino acids.
Conflict of interest statement
Figures

Similar articles
-
Genome-Wide Analysis of Domain-Swap Predicted Products in the Genome of Anti-Stress Medicinal Plant: Ocimum tenuiflorum.Bioinform Biol Insights. 2019 Jan 9;13:1177932218821362. doi: 10.1177/1177932218821362. eCollection 2019. Bioinform Biol Insights. 2019. PMID: 30692846 Free PMC article.
-
Functional repertoire, molecular pathways and diseases associated with 3D domain swapping in the human proteome.J Clin Bioinforma. 2012 Apr 3;2(1):8. doi: 10.1186/2043-9113-2-8. J Clin Bioinforma. 2012. PMID: 22472218 Free PMC article.
-
Insights into Protein Sequence and Structure-Derived Features Mediating 3D Domain Swapping Mechanism using Support Vector Machine Based Approach.Bioinform Biol Insights. 2010 Jun 17;4:33-42. doi: 10.4137/bbi.s4464. Bioinform Biol Insights. 2010. PMID: 20634983 Free PMC article.
-
3D domain swapping: as domains continue to swap.Protein Sci. 2002 Jun;11(6):1285-99. doi: 10.1110/ps.0201402. Protein Sci. 2002. PMID: 12021428 Free PMC article. Review.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
-
A cardiologist's guide to machine learning in cardiovascular disease prognosis prediction.Basic Res Cardiol. 2023 Mar 20;118(1):10. doi: 10.1007/s00395-023-00982-7. Basic Res Cardiol. 2023. PMID: 36939941 Free PMC article. Review.
-
Genome-Wide Analysis of Domain-Swap Predicted Products in the Genome of Anti-Stress Medicinal Plant: Ocimum tenuiflorum.Bioinform Biol Insights. 2019 Jan 9;13:1177932218821362. doi: 10.1177/1177932218821362. eCollection 2019. Bioinform Biol Insights. 2019. PMID: 30692846 Free PMC article.
-
Exploring the Roles of Proline in Three-Dimensional Domain Swapping from Structure Analysis and Molecular Dynamics Simulations.Protein J. 2018 Feb;37(1):13-20. doi: 10.1007/s10930-017-9747-5. Protein J. 2018. PMID: 29119487
References
-
- Crestfield AM, Stein WH, Moor S. On the aggregation of bovine pancreatic ribonuclease. Arch Biochem Biophys. 1962. September;Suppl 1:217–22. - PubMed
-
- Gordon-Smith DJ, Carbajo RJ, Stott K, Neuhaus D. Solution studies of chymotrypsin inhibitor-2 glutamine insertion mutants show no interglutamine interactions. Biochem Biophys Res Commun. 2001. January 26;280(3):855–60. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

