WALT: fast and accurate read mapping for bisulfite sequencing
- PMID: 27466624
- PMCID: PMC5181568
- DOI: 10.1093/bioinformatics/btw490
WALT: fast and accurate read mapping for bisulfite sequencing
Abstract
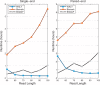
Whole-genome bisulfite sequencing (WGBS) has emerged as the gold-standard technique in genome-scale studies of DNA methylation. Mapping reads from WGBS requires unique considerations that make the process more time-consuming than in other sequencing applications. Typical WGBS data sets contain several hundred million reads, adding to this analysis challenge. We present the WALT tool for mapping WGBS reads. WALT uses a strategy of hashing periodic spaced seeds, which leads to significant speedup compared with the most efficient methods currently available. Although many existing WGBS mappers slow down with read length, WALT improves in speed. Importantly, these speed gains do not sacrifice accuracy.
Availability and implementation: WALT is available under the GPL v3 license, and downloadable from https://github.com/smithlabcode/walt.
Contact: andrewds@usc.edu or tingchen@usc.edu SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
An integrative approach for efficient analysis of whole genome bisulfite sequencing data.BMC Genomics. 2015;16 Suppl 12(Suppl 12):S14. doi: 10.1186/1471-2164-16-S12-S14. Epub 2015 Dec 9. BMC Genomics. 2015. PMID: 26680746 Free PMC article.
-
Evaluation of preprocessing, mapping and postprocessing algorithms for analyzing whole genome bisulfite sequencing data.Brief Bioinform. 2016 Nov;17(6):938-952. doi: 10.1093/bib/bbv103. Epub 2015 Dec 1. Brief Bioinform. 2016. PMID: 26628557 Free PMC article.
-
MethylStar: A fast and robust pre-processing pipeline for bulk or single-cell whole-genome bisulfite sequencing data.BMC Genomics. 2020 Jul 13;21(1):479. doi: 10.1186/s12864-020-06886-3. BMC Genomics. 2020. PMID: 32660416 Free PMC article.
-
Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing.BMC Genomics. 2017 Jan 5;18(1):31. doi: 10.1186/s12864-016-3392-9. BMC Genomics. 2017. PMID: 28056787 Free PMC article.
-
Efficiently quantifying DNA methylation for bulk- and single-cell bisulfite data.Bioinformatics. 2023 Jun 1;39(6):btad386. doi: 10.1093/bioinformatics/btad386. Bioinformatics. 2023. PMID: 37326968 Free PMC article.
Cited by
-
The Australian dingo is an early offshoot of modern breed dogs.Sci Adv. 2022 Apr 22;8(16):eabm5944. doi: 10.1126/sciadv.abm5944. Epub 2022 Apr 22. Sci Adv. 2022. PMID: 35452284 Free PMC article.
-
Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity.Dev Cell. 2019 Jan 7;48(1):49-63.e7. doi: 10.1016/j.devcel.2018.11.048. Dev Cell. 2019. PMID: 30620902 Free PMC article.
-
Human TSC2 Mutant Cells Exhibit Aberrations in Early Neurodevelopment Accompanied by Changes in the DNA Methylome.bioRxiv [Preprint]. 2024 Jun 6:2024.06.04.597443. doi: 10.1101/2024.06.04.597443. bioRxiv. 2024. PMID: 38895266 Free PMC article. Preprint.
-
Chromosome-length genome assembly and structural variations of the primal Basenji dog (Canis lupus familiaris) genome.BMC Genomics. 2021 Mar 16;22(1):188. doi: 10.1186/s12864-021-07493-6. BMC Genomics. 2021. PMID: 33726677 Free PMC article.
-
Strong Parallel Differential Gene Expression Induced by Hatchery Rearing Weakly Associated with Methylation Signals in Adult Coho Salmon (O. kisutch).Genome Biol Evol. 2022 Apr 10;14(4):evac036. doi: 10.1093/gbe/evac036. Genome Biol Evol. 2022. PMID: 35276004 Free PMC article.
References
-
- Bock C. (2012) Analysing and interpreting DNA methylation data. Nat. Rev. Gen., 13, 705–719. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources