Stress Physiology of Lactic Acid Bacteria
- PMID: 27466284
- PMCID: PMC4981675
- DOI: 10.1128/MMBR.00076-15
Stress Physiology of Lactic Acid Bacteria
Abstract
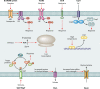
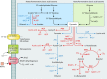
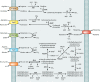
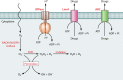
Lactic acid bacteria (LAB) are important starter, commensal, or pathogenic microorganisms. The stress physiology of LAB has been studied in depth for over 2 decades, fueled mostly by the technological implications of LAB robustness in the food industry. Survival of probiotic LAB in the host and the potential relatedness of LAB virulence to their stress resilience have intensified interest in the field. Thus, a wealth of information concerning stress responses exists today for strains as diverse as starter (e.g., Lactococcus lactis), probiotic (e.g., several Lactobacillus spp.), and pathogenic (e.g., Enterococcus and Streptococcus spp.) LAB. Here we present the state of the art for LAB stress behavior. We describe the multitude of stresses that LAB are confronted with, and we present the experimental context used to study the stress responses of LAB, focusing on adaptation, habituation, and cross-protection as well as on self-induced multistress resistance in stationary phase, biofilms, and dormancy. We also consider stress responses at the population and single-cell levels. Subsequently, we concentrate on the stress defense mechanisms that have been reported to date, grouping them according to their direct participation in preserving cell energy, defending macromolecules, and protecting the cell envelope. Stress-induced responses of probiotic LAB and commensal/pathogenic LAB are highlighted separately due to the complexity of the peculiar multistress conditions to which these bacteria are subjected in their hosts. Induction of prophages under environmental stresses is then discussed. Finally, we present systems-based strategies to characterize the "stressome" of LAB and to engineer new food-related and probiotic LAB with improved stress tolerance.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

















Similar articles
-
Stress responses in lactic acid bacteria.Antonie Van Leeuwenhoek. 2002 Aug;82(1-4):187-216. Antonie Van Leeuwenhoek. 2002. PMID: 12369188 Review.
-
Interactions among lactic acid starter and probiotic bacteria used for fermented dairy products.J Dairy Sci. 2002 Apr;85(4):721-9. doi: 10.3168/jds.S0022-0302(02)74129-5. J Dairy Sci. 2002. PMID: 12018416
-
A large factory-scale application of selected autochthonous lactic acid bacteria for PDO Pecorino Siciliano cheese production.Food Microbiol. 2016 Oct;59:66-75. doi: 10.1016/j.fm.2016.05.011. Epub 2016 May 18. Food Microbiol. 2016. PMID: 27375245
-
In vitro assessment of functional properties of lactic acid bacteria isolated from faecal microbiota of healthy dogs for potential use as probiotics.Benef Microbes. 2013 Sep;4(3):267-75. doi: 10.3920/BM2012.0048. Benef Microbes. 2013. PMID: 23538205
-
Mechanisms and improvement of acid resistance in lactic acid bacteria.Arch Microbiol. 2018 Mar;200(2):195-201. doi: 10.1007/s00203-017-1446-2. Epub 2017 Oct 26. Arch Microbiol. 2018. PMID: 29075866 Review.
Cited by
-
Identification of the effects of acid-resistant Lactobacillus casei metallopeptidase gene under colon-specific promoter on the colorectal and breast cancer cell lines.Iran J Basic Med Sci. 2021 Apr;24(4):506-513. doi: 10.22038/ijbms.2021.53015.11950. Iran J Basic Med Sci. 2021. PMID: 34094033 Free PMC article.
-
Engineering Polymeric Nanosystems against Oral Diseases.Molecules. 2021 Apr 13;26(8):2229. doi: 10.3390/molecules26082229. Molecules. 2021. PMID: 33924289 Free PMC article. Review.
-
Influence of nitrogen sources on the tolerance of Lacticaseibacillus rhamnosus to heat stress and oxidative stress.J Ind Microbiol Biotechnol. 2022 Oct 13;49(5):kuac020. doi: 10.1093/jimb/kuac020. J Ind Microbiol Biotechnol. 2022. PMID: 36073749 Free PMC article.
-
Negative chemotaxis of Ligilactobacillus agilis BKN88 against gut-derived substances.Sci Rep. 2023 Sep 20;13(1):15632. doi: 10.1038/s41598-023-42840-5. Sci Rep. 2023. PMID: 37730901 Free PMC article.
-
Genetics of Lactococci.Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.gpp3-0035-2018. doi: 10.1128/microbiolspec.GPP3-0035-2018. Microbiol Spectr. 2019. PMID: 31298208 Free PMC article. Review.
References
-
- Papadimitriou K, Pot B, Tsakalidou E. 2015. How microbes adapt to a diversity of food niches. Curr Opin Food Sci 2:29–35. doi:10.1016/j.cofs.2015.01.001. - DOI
-
- Orla-Jensen S. 1919. The lactic acid bacteria. Fred Host and Son, Copenhagen, Denmark.
-
- Sun Z, Harris HM, McCann A, Guo C, Argimon S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O'Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O'Toole PW. 2015. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322. doi:10.1038/ncomms9322. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

