Structure of the adenosine A(2A) receptor bound to an engineered G protein
- PMID: 27462812
- PMCID: PMC4979997
- DOI: 10.1038/nature18966
Structure of the adenosine A(2A) receptor bound to an engineered G protein
Erratum in
-
Erratum: Structure of the adenosine A2A receptor bound to an engineered G protein.Nature. 2016 Oct 27;538(7626):542. doi: 10.1038/nature19803. Epub 2016 Sep 14. Nature. 2016. PMID: 27629518 No abstract available.
Abstract
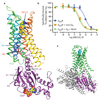
G-protein-coupled receptors (GPCRs) are essential components of the signalling network throughout the body. To understand the molecular mechanism of G-protein-mediated signalling, solved structures of receptors in inactive conformations and in the active conformation coupled to a G protein are necessary. Here we present the structure of the adenosine A(2A) receptor (A(2A)R) bound to an engineered G protein, mini-Gs, at 3.4 Å resolution. Mini-Gs binds to A(2A)R through an extensive interface (1,048 Å2) that is similar, but not identical, to the interface between Gs and the β2-adrenergic receptor. The transition of the receptor from an agonist-bound active-intermediate state to an active G-protein-bound state is characterized by a 14 Å shift of the cytoplasmic end of transmembrane helix 6 (H6) away from the receptor core, slight changes in the positions of the cytoplasmic ends of H5 and H7 and rotamer changes of the amino acid side chains Arg3.50, Tyr5.58 and Tyr7.53. There are no substantial differences in the extracellular half of the receptor around the ligand binding pocket. The A(2A)R-mini-Gs structure highlights both the diversity and similarity in G-protein coupling to GPCRs and hints at the potential complexity of the molecular basis for G-protein specificity.
Figures


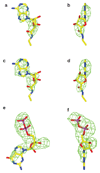



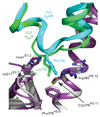
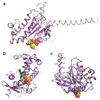
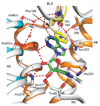



Similar articles
-
Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation.Nature. 2011 May 18;474(7352):521-5. doi: 10.1038/nature10136. Nature. 2011. PMID: 21593763 Free PMC article.
-
Structure of an agonist-bound human A2A adenosine receptor.Science. 2011 Apr 15;332(6027):322-7. doi: 10.1126/science.1202793. Epub 2011 Mar 10. Science. 2011. PMID: 21393508 Free PMC article.
-
Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein.Elife. 2018 May 4;7:e35946. doi: 10.7554/eLife.35946. Elife. 2018. PMID: 29726815 Free PMC article.
-
Agonist-bound structures of G protein-coupled receptors.Curr Opin Struct Biol. 2012 Aug;22(4):482-90. doi: 10.1016/j.sbi.2012.03.007. Epub 2012 Apr 3. Curr Opin Struct Biol. 2012. PMID: 22480933 Review.
-
Structural insights into agonist-induced activation of G-protein-coupled receptors.Curr Opin Struct Biol. 2011 Aug;21(4):541-51. doi: 10.1016/j.sbi.2011.06.002. Epub 2011 Jun 30. Curr Opin Struct Biol. 2011. PMID: 21723721 Review.
Cited by
-
Discovery and Structure-Activity Relationship Studies of Novel Adenosine A1 Receptor-Selective Agonists.J Med Chem. 2022 Nov 10;65(21):14864-14890. doi: 10.1021/acs.jmedchem.2c01414. Epub 2022 Oct 21. J Med Chem. 2022. PMID: 36270633 Free PMC article.
-
Ligand modulation of the conformational dynamics of the A2A adenosine receptor revealed by single-molecule fluorescence.Sci Rep. 2021 Mar 15;11(1):5910. doi: 10.1038/s41598-021-84069-0. Sci Rep. 2021. PMID: 33723285 Free PMC article.
-
Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.Proc Natl Acad Sci U S A. 2019 Aug 13;116(33):16394-16403. doi: 10.1073/pnas.1906658116. Epub 2019 Jul 30. Proc Natl Acad Sci U S A. 2019. PMID: 31363053 Free PMC article.
-
Gs protein peptidomimetics as allosteric modulators of the β2-adrenergic receptor.RSC Adv. 2018 Jan 9;8(4):2219-2228. doi: 10.1039/c7ra11713b. eCollection 2018 Jan 5. RSC Adv. 2018. PMID: 35542596 Free PMC article.
-
Network-driven intracellular cAMP coordinates circadian rhythm in the suprachiasmatic nucleus.Sci Adv. 2023 Jan 4;9(1):eabq7032. doi: 10.1126/sciadv.abq7032. Epub 2023 Jan 4. Sci Adv. 2023. PMID: 36598978 Free PMC article.
References
-
- Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

