The Chp1 chromodomain binds the H3K9me tail and the nucleosome core to assemble heterochromatin
- PMID: 27462451
- PMCID: PMC4849473
- DOI: 10.1038/celldisc.2016.4
The Chp1 chromodomain binds the H3K9me tail and the nucleosome core to assemble heterochromatin
Abstract
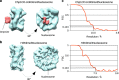
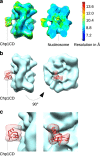
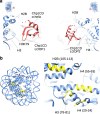
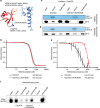
To maintain genome stability, cells pack large portions of their genome into silent chromatin or heterochromatin. Histone H3 lysine 9 methylation, a hallmark of heterochromatin, is recognized by conserved readers called chromodomains. But how chromodomains interact with their actual binding partner, the H3K9 methylated nucleosome, remains elusive. We have determined the structure of a nucleosome trimethylated at lysine 9 of histone H3 (H3K9me3 Nucleosome) in a complex with the chromodomain of Chp1, a protein required for RNA interference-dependent heterochromatin formation in fission yeast. The cryo-electron microscopy structure reveals that the chromodomain of Chp1 binds the histone H3 lysine 9 methylated tail and the core of the nucleosome, primarily histones H3 and H2B. Mutations in chromodomain of Chp1 loops, which interact with the nucleosome core, abolished this interaction in vitro. Moreover, fission yeast cells with Chp1 loop mutations have a defect in Chp1 recruitment and heterochromatin formation. This study reveals the structural basis for heterochromatic silencing and suggests that chromodomains could read histone code in the H3 tail and the nucleosome core, which would provide an additional layer of regulation.
Keywords: Chp 1; RNAi; S. pombe; chromodomain; cryo-EM; heterochromatin; nucleosome.
Figures
Similar articles
-
RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast.Mol Cell Biol. 2005 Mar;25(6):2331-46. doi: 10.1128/MCB.25.6.2331-2346.2005. Mol Cell Biol. 2005. PMID: 15743828 Free PMC article.
-
Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing.Mol Cell. 2012 Jul 27;47(2):228-41. doi: 10.1016/j.molcel.2012.05.017. Epub 2012 Jun 21. Mol Cell. 2012. PMID: 22727667
-
cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site.Curr Biol. 2002 Oct 1;12(19):1652-60. doi: 10.1016/s0960-9822(02)01177-6. Curr Biol. 2002. PMID: 12361567
-
Phosphorylation of repressive histone code readers by casein kinase 2 plays diverse roles in heterochromatin regulation.J Biochem. 2019 Jul 1;166(1):3-6. doi: 10.1093/jb/mvz045. J Biochem. 2019. PMID: 31198932 Review.
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
Cited by
-
Ccr4-Not complex reduces transcription efficiency in heterochromatin.Nucleic Acids Res. 2022 Jun 10;50(10):5565-5576. doi: 10.1093/nar/gkac403. Nucleic Acids Res. 2022. PMID: 35640578 Free PMC article.
-
Disordered region of H3K9 methyltransferase Clr4 binds the nucleosome and contributes to its activity.Nucleic Acids Res. 2019 Jul 26;47(13):6726-6736. doi: 10.1093/nar/gkz480. Nucleic Acids Res. 2019. PMID: 31165882 Free PMC article.
-
Tailing and degradation of Argonaute-bound small RNAs protect the genome from uncontrolled RNAi.Nat Commun. 2017 May 25;8:15332. doi: 10.1038/ncomms15332. Nat Commun. 2017. PMID: 28541282 Free PMC article.
-
Cryo-electron microscopy of chromatin biology.Acta Crystallogr D Struct Biol. 2017 Jun 1;73(Pt 6):541-548. doi: 10.1107/S2059798317004430. Epub 2017 Apr 20. Acta Crystallogr D Struct Biol. 2017. PMID: 28580916 Free PMC article. Review.
-
Microscale Thermophoresis Analysis of Chromatin Interactions.Methods Mol Biol. 2024;2819:357-379. doi: 10.1007/978-1-0716-3930-6_17. Methods Mol Biol. 2024. PMID: 39028515
References
-
- Bannister AJ , Zegerman P , Partridge JF et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001; 410: 120–124. - PubMed
-
- Lachner M , O’Carroll D , Rea S , Mechtler K , Jenuwein T . Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410: 116–120. - PubMed
-
- Nakayama J , Rice JC , Strahl BD , Allis CD , Grewal SI . Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 2001; 292: 110–113. - PubMed
-
- Rea S , Eisenhaber F , O’Carroll D et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406: 593–599. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

